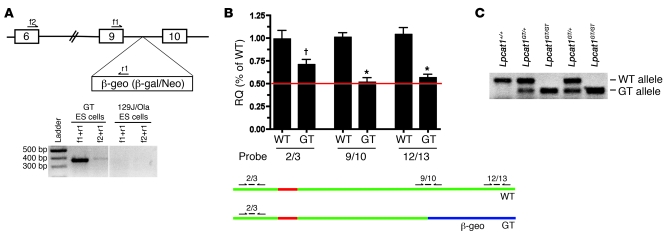
Figure 4. Confirmation of Lpcat1 GT in ES cells and generation of mice.
(A) Top: Diagram of Lpcat1 locus. Boxes denote exons, and intervening lines denote introns. The β-geo selection cassette encoding a β-galactosidase/neomycin fusion protein was inserted in intron 9, approximately 400 bp 3′ of exon 9. f1, f2, and r2 denote primers used to confirm the presence of GT cassette in the Lpcat1 locus by qPCR analysis. Bottom: PCR analysis of the Lpcat1 locus in GT ES cells or parental ES cells (129J/Ola) using 2 separate primer sets. The presence of amplicons in GT ES cells with the absence of signal in parental cells demonstrates trapping of the Lpcat1 locus at exon 9. Vertical line indicates discontinuous lanes in the same gel. (B) qPCR analysis of GT ES cells. cDNA from GT ES cells (GT) and parental cells (WT) was subjected to qPCR analysis using 3 separate exon-spanning TaqMan primer/probe sets for Lpcat1 as depicted in the bottom panel: 2/3, 9/10, and 12/13. Data were normalized to Actb and represent 3 independent experiments, each performed in triplicate. Note the difference in signal between 2/3 probe and 9/10 probe in GT cells. †P < 0.05 versus 9/10 and 12/13 GT;*P < 0.05 versus 2/3 WT. RQ, relative quantification. (C) Multiplex PCR genotyping of genomic DNA from progeny of Lpcat1GT/+ intercrosses. Amplicon for WT allele is approximately 350 bp; GT, approximately 300 bp.