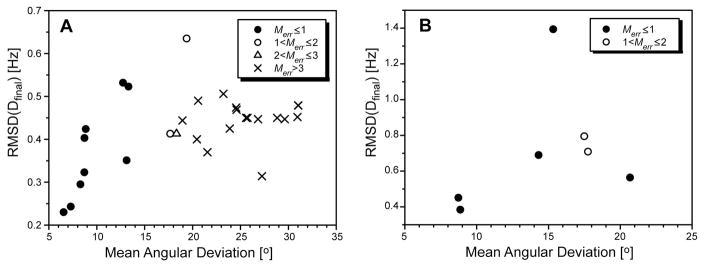
Figure 6.
Plot of all unique solutions resulting from application of the RSDC protocol to the first 3 OLC-RDCs of ubiquitin (A) and GB1 (B). The RMSD between the experimental RDCs and those determined by RSDC is utilized to select the best solution under the restraint that optimal tensor magnitude be maintained (0 ≤ Merr ≤ 1). The structural accuracy is reported as the mean angular deviation for each RSDC solution from the relevant X-ray structures for ubiquitin (1UBQ) (Vijaykumar et al. 1987) and protein GB1 (1PGB) (Gallagher et al. 1994).