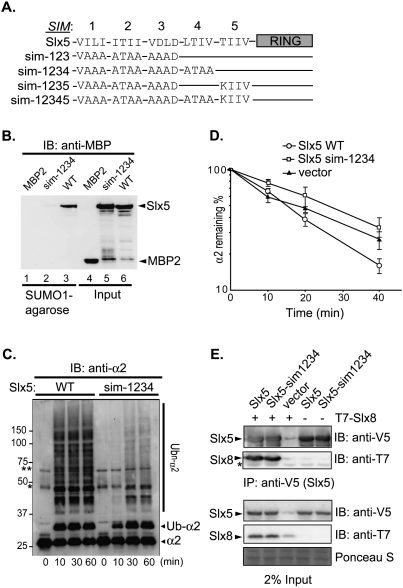
Figure 6.
The SIMs of Slx5 are required for SUMO binding and α2 degradation. (A) Schematic of Slx5 (wild type and mutants) with its SIMs and RING domain highlighted. Four residues that constitute the hydrophobic core of each SIM are indicated, with the altered sequences indicated for each mutant. The SIM core sequences that were tested were SIM-1, 24VILI27; SIM-2, 93ITII96; SIM-3, 116VDLD119; SIM-4, 155LTIV158; and SIM-5, 476TIIV479. (B) Protein binding to SUMO1-agarose. (Lanes 1–3) Purified recombinant MBP-Slx5 (wild type and sim-1234) or MBP2 proteins were incubated with SUMO1-agarose, and proteins that bound to the resin were eluted, resolved by SDS-PAGE, and detected by anti-MBP immunoblotting. Input proteins (20%) are shown in lanes 4–6. (C) Slx5–Slx8-mediated ubiquitylation of α2 in vitro (conditions as in Fig. 3). Reactions were stopped at the indicated times by addition of 2× SDS gel loading buffer. Proteins were resolved by SDS-PAGE, and were detected by anti-α2 immunoblotting. Asterisks denote the same two cross-reacting proteins noted in Figure 3. (D) Quantitation of α2 degradation in doa10Δ slx5Δ strains transformed with plasmid-borne SLX5 alleles measured by pulse-chase analysis at 30°C. (E) Coimmunoprecipitation of T7-tagged Slx8 with variants of Slx5. Analysis was done as in Figure 5. Slx5 variants were detected by anti-V5 immunoblotting, and coprecipitated T7-Slx8 was detected by anti-T7 immunoblotting. Protein staining of a portion of the membrane shows similar loading.