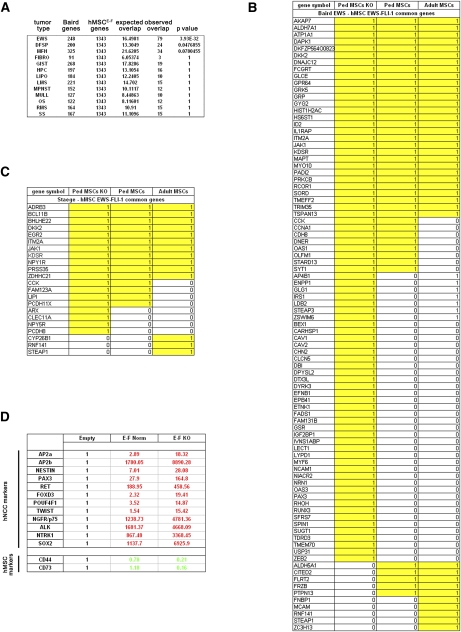
Figure 3.
hpMSCs cultured in KO stem cell culture conditions display increased permissiveness for EWS-FLI-1 activity and undergo genetic reprogramming to acquire a NCSC molecular phenotype. (A) Statistical analysis of the genes common to hpMSCEWS-FLI-1s and the soft tissue tumor gene expression profile database (Baird et al. 2005) presented in Figure 2, showing the marked increase in similarity between hpMSCEWS-FLI-1s and ESFTs when hpMSCs were cultured in KO conditions. (B) List of the 79 genes found to be shared by hpMSCEWS-FLI-1s grown in KO culture conditions and the ESFT gene expression profile, and their comparison with the genes induced in pediatric and adult hMSCEWS-FLI-1s cultured in presence of serum. (C) List of the 19 genes common to hpMSCEWS-FLI-1s cultured in KO conditions and the ESFT expression signature identified by Staege et al. (2004) compared with results obtained using pediatric and adult MSCs cultured in the presence of serum. (D) Expression levels of a selected panel of transcripts, currently used to define the NCSC phenotype, as assessed by real-time PCR, showing induction of all of the genes in hpMSCEWS-FLI-1s cultured in KO medium but not in serum-supplemented medium. In KO medium-cultured cells only, induction of hNCSC transcripts was accompanied by a concomitant repression of the two major MSC markers, CD73 and CD44. Real-time PCR experiments were normalized to cyclophyllin A and done in triplicate. Error bars represent the SD of three independent determinations. For abbreviations, see the legend for Figure 2.