Figure 2.
Genomic Organization and Similarity of the GER4 Genes.
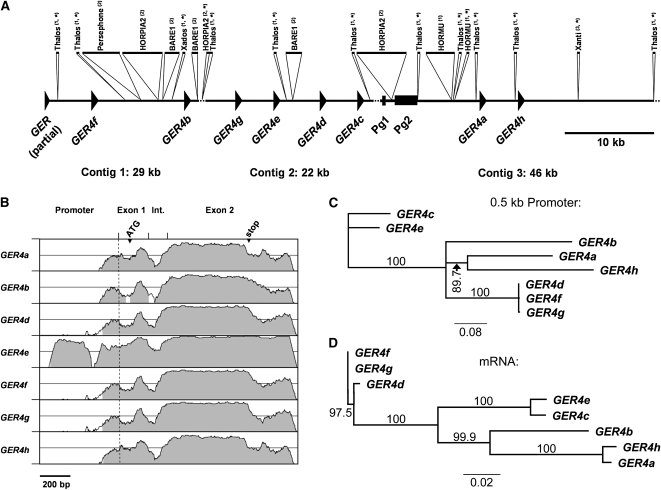
(A) Annotation of genes and repetitive elements on sequenced barley BAC clone 418E01. The three assembled contigs of 29, 22, and 46 kb are represented schematically. The annotated coding sequences of the GER subfamily 4 (GER4a to GER4 h and a partial GER gene) are shown as arrowheads (exon/intron structure not shown). Two pseudogenes are indicated as boxes (Pg1, fragment of a germin-like gene; Pg2, sequence with homology to phospholipase D). The sequence contains several repetitive elements that are indicated as inserts above: (1) DNA transposons, (2) retrotransposons, and (3) other. The sequences are drawn to scale except for elements that otherwise are too small to be represented appropriately (asterisk). Stippled lines indicate remaining short sequence gaps, which prevented the determination of the orientation of the central contig 2, and a sequence stretch extending toward the right border of the BAC that did not contain any annotated gene or element.
(B) Similarity of GER4a to GER4 h genes. The genomic sequences comprising 600-bp promoter region, two exons, and the intron (Int.) of GER4a to GER4 h are represented in a VISTA plot using the GER4c sequence as a reference (x axis, window size 100 bp). The similarities between the different GER4 genes and GER4c are plotted on the y axis each spanning the range between 50 and 100%. The position of the transcriptional start (stippled line), translational start ATG, and stop codon is indicated.
(C) Unrooted neighbor-joining tree of GER4 proximal promoter sequences. The bar indicates 8% sequence dissimilarity. Bootstrap values (in percentage) are indicated.
(D) Unrooted neighbor-joining tree of GER4 mRNA sequences. The bar indicates 2% sequence dissimilarity. Bootstrap values (in percentage) are indicated.