Fig. 1. Successive assembly of a hierarchical network identifies ISV in Tg(fli1:EGFP)y1 embryos.
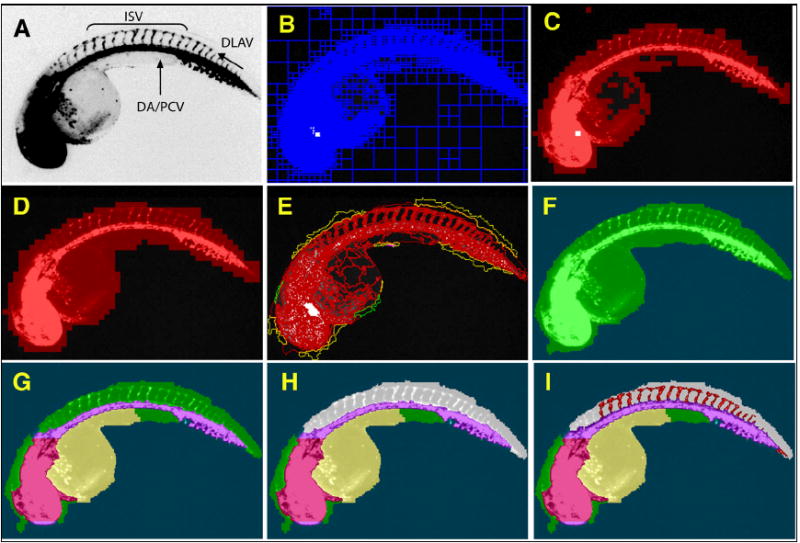
48 hpf Tg(fli1:EGFP)y1 zebrafish embryos were imaged on an ArrayScan II high-content reader equipped with a 1.25x objective. The original image (A) was segmented based on pixel intensities and regional variability (B). Regions of high variability (C) were fused and expanded to provide a general outline (D). The outline was refined (E) to demarcate the whole zebrafish embryo (F). Specific subdomains within the embryo such as head, dorsal aorta/posterior cardinal vein, yolk (G), and dorsal area (H, grey) were then assigned through successive interactive loops of locally specific segmentation and classification, resulting in a hierarchical structure of the entire embryo with its subdomains. Knowledge generated during each prior classification step enabled the ruleset to specifically quantify ISV in the dorsal tail (I, red) without interference from other fluorescent regions. (DA), Dorsal aorta; (PCV), posterior cardinal vein; (ISV), intersegmental vessels; and (DLAV), dorsal longitudinal anastomotic vessel.
