Abstract
A method to perform in-situ roughening of arrays of microstructures weakly adherent to an underlying substrate was presented. SU8, 1002F, and polydimethylsiloxane (PDMS) microstructures were roughened by polishing with a particle slurry. The roughness and the percentage of dislodged or damaged microstructures was evaluated as a function of the roughening time for both SU8 and 1002F structures. A maximal RMS roughness of 7-18 nm for the surfaces was obtained within 15 to 30 s of polishing with the slurry. This represented a 4-9 fold increase in surface roughness relative to that of the native surface. Less than 0.8% of the microstructures on the array were removed or damage after 5 min of polishing. Native and roughened arrays were assessed for their ability to support fibronectin adhesion and cell attachment and growth. The quantity of adherent fibronectin was increased on roughened arrays by two-fold over that on native arrays. Cell adhesion to the roughened surfaces was also increased compared to native surfaces. Surface roughening with the particle slurry also improved the ability to stamp molecules onto the substrate during microcontact printing. Roughening both the PDMS stamp and substrate resulted in up to a 20-fold improvement in the transfer of BSA-Alexa Fluor 647 from the stamp to the substrate. Thus roughening of micron-scale surfaces with a particle slurry increased the adhesion of biomolecules as well as cells to microstructures with little to no damage to large scale arrays of the structures.
Keywords: Roughening, in-situ, polymer, MEMS, microarray, printing, cell
INTRODUCTION
The need for miniaturization is especially apparent in biochemical and biomedical analysis since smaller scale devices provide better efficiency, integration, and throughput. Microscale devices can also permit automation, minimize sample and reagent consumption, speed up analysis, and improve system portability (1, 2). More recently, polymers have found increased usage in nearly all types of microelectromechanical systems (MEMS) (2). While early studies focused primarily on silicon- and glass-based devices, polymers have attractive features such as biocompatibility, low cost, and simple fabrication methods (2, 3). In addition, unlike the limited choices provided by glass or silicon substrates, there are usually numerous options from a pool of available polymeric substrate materials for a specific MEMS applications (2, 4). Many polymeric devices have focused on the analysis of biomolecules such as oligonucleotides, peptides and proteins (5, 6). However, analysis systems utilizing cultured cells on a microchip platform are rapidly growing (1). These applications include cell culture, separation, lysis, fixation, and immunostaining (1, 7, 8). In most cases, the surface properties of a polymeric microdevice must be modified to match the specific cell-based application of the device (2-4). Surface modification has been accomplished by covalently and noncovalently attached chemical coatings as well as other strategies for example by bombardment with ions. However each of these technologies significantly alters the chemical moieties present on the surface of the microdevice.
It is often of interest to alter the surface of a microdevice without changing the chemical composition of the surface. For example, the physical roughening of a surface can improve wettability as well as adhesion to another surface without the need for chemical alterations. Roughening of a flat surface or wafer is often accomplished using slurries of particles (9-14). These technologies often require very expensive mechanical, chemical, or electrochemical instrumentation and will frequently damage delicate features or remove loosely attached microstructures from their substrate. To improve surface roughness, the use of complicated and expensive nanotechnology-based techniques to fabricate nanostructures varied in shape, type and dimension has been reported (15-28). The nanotechnology-based studies were predominantly directed at flat surfaces and usually cannot be transferred to miniaturized 3-D features without challenges.
In this work, we report a simple, rapid and inexpensive method using a slurry to roughen polymeric microstructures formed from two photoresists or PDMS. Surface roughness was characterized by atomic force microscopy (AFM) with respect to the duration of the roughening process. In addition, fragmentation and loss of weakly attached microstructures from an array was also evaluated. Chemical alterations in the roughened surfaces were assessed utilizing X-ray photoelectron spectroscopy (XPS). The influence of the roughening strategy on the quantity of fibronectin and the numbers of cells attached to the surface were measured. The utility of this roughening method was demonstrated in cell separation by micropallet arrays and for microcontact printing (μCP) of biomolecules as two examples of microdevices utilized in biomedical research. The ability of cells to remain adherent to a growth surface during sorting as well as the efficiency of ink transfer to a surface during μCP was assessed.
EXPERIMENTAL SECTION
Materials
EPON resin 1002F (phenol, 4,4'-(1-methylethylidene)bis-, polymer with 2,2'-[(1-methylethylidene) bis(4,1-phenyleneoxymethylene]bis[oxirane]) was obtained from Miller-Stephenson (Sylmar, CA). SU8 photoresist (a polymeric solid epoxy novolac resin possessing an average epoxide group functionality around eight) and SU8 developer (1-methoxy-2-propyl acetate, also used for 1002F) were obtained from MicroChem Corp. (Newton, MA). The Sylgard 184 silicone elastomer kit used to form poly(dimethyl siloxane) (PDMS) components was purchased from Dow Corning (Midland, MI). (Heptadecafluoro-1,1,2,2-tetrahydrodecyl) trichlorosilane was from Gelest Inc. (Morrisville, PA). Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum (FBS), L-glutamine, penicillin/streptomycin, phosphate buffered saline (PBS) 1x, pH 7.4, 0.05% trypsin with EDTA solution, Alexa Fluor 647 labeled bovine serum albumin (BSA-Alexa 647, excitation 650 nm, emission 668 nm), and Alexa Fluor 633 protein labeling kit (excitation 632 nm, emission 647 nm) were obtained from Invitrogen (Carlsbad, CA). Human plasma fibronectin was purchased from Millipore Corporation (Billerica, MA). Slurry of alumina particles (1.0 μm) in water was purchased from MTI Corp. (Richmond, CA). All other chemicals were obtained from Fisher Scientific (Pittsburgh, PA).
Fabrication of SU8 and 1002F Micropallet Arrays
Micropallet arrays composed of SU8 or 1002F (50 μm sides, 20 μm inter-pallet spacing, and 50 μm height) were fabricated on a glass substrate as described previously (29, 30). O-rings (I.D./O.D. of 18/35 mm) to form chambers around the arrays were constructed from PDMS as described previously (29, 30). O-rings were placed on a micropallet array and then uncured PDMS applied around the O-ring edges followed by curing of the PDMS (65°C, 30 min). The SU8 and 1002F micropallet arrays with attached cell chamber contained approximately 46,000 micropallets.
Surface Roughening with Alumina Particles
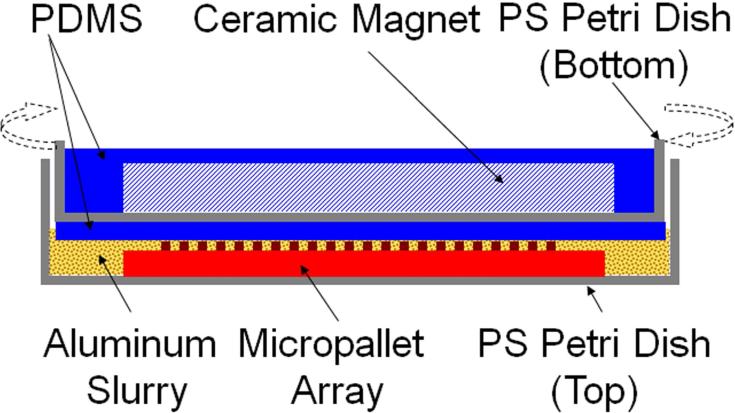
A custom-fabricated device utilized an aqueous slurry of alumina particles to roughen the pallet surfaces on the arrays, the PDMS stamps, and photoresist films (Figures 1 & S1). The top portion of the device was fabricated in the lower portion of a polystyrene tissue culture dish (60 × 15 mm, Corning Incorporated, Corning, NY). A ceramic magnet (oval shaped, 38 mm diameter-major axis, 8 mm diameter-minor axis, Fisher Scientific) was sealed within a layer of PDMS inside the lower portion of the polystyrene tissue culture dish. Another layer of PDMS covered the portion of the culture dish in contact with the particle slurry. The bottom piece of the polishing device was the upper part (lid) of the polystyrene tissue culture dish. A self-adhering polishing cloth was placed inside this component to prevent the movement of the microarray, PDMS stamp, or photoresist film during roughening. After each use, the bottom and top pieces of the roughening device were washed with water and ethanol.
FIGURE 1.
Schematic side view of the custom device used for roughening of micropallet arrays, PDMS stamps, and photoresist films. A micropallet array is shown in the device which is fabricated from a standard polystyrene dish.
To roughen their surface, a micropallet array, PDMS stamp, or photoresist film was placed in the bottom piece of the roughening device and overlaid with a 1:1 (v/v) solution (5 mL) of alumina particles in distilled water. The bottom piece was then placed on a magnetic stirrer (PC-620 Corning Incorporated, Corning, NY). The top piece of the device was placed over the bottom piece and rotated at 66 rpm for 15 s (SU8) or 30 s (1002F) unless otherwise indicated. The roughened micropallet arrays were rinsed with distilled water and then ethanol five times and dried with N2. For cell-culture experiments only, the roughened and unroughened arrays were coated with heptadecafluoro-1,1,2,2-tetrahydrodecyl trichlorosilane in a low-pressure reactor (31). The micropallet arrays were stored in a vacuum desiccator until use.
Surface Analyses
Contact-mode AFM images of 1002F micropallets were obtained in air using a Molecular Force Probe 3D AFM (Asylum Research, Santa Barbara, CA) controlled with MFP-3D software running under Igor Pro (Wavemetrics, Lake Oswego, OR). A ThermoMicroscopes AFM from Topometrix (Santa Clara, CA) controlled with SPM Image software (Nanoscience Instruments Inc., Phoenix, AZ) was used for obtaining contact mode AFM images of SU8 micropallets in air. The AFM data for SU8 were plotted and analyzed using Gwyddion software (Czech Metrology Institute, Brno, Czech Republic). The root mean squared (RMS) roughness was used to report the roughness values (32). Each AFM measurement was obtained on randomly selected 25-μm2 regions on the top surfaces of the pallets.
XPS was used for surface chemical analysis of polymers before and after roughening. All XPS data were acquired on a Kratos Axis Ultra DLD system (Kratos Analytical Inc., New York, NY) with a monochromatic Al Ka source. Survey scans and high resolution scans were acquired at pass energies of 80 eV and 20 eV, respectively. All spectra were calibrated using the C1s energy of 284.6 eV. The high resolution scans were background subtracted. Sessile water contact angle measurements were performed using a method described previously (4).
Coating SU8 and 1002F Micropallet Arrays with Alexa Fluor 633-Labeled Fibronectin
For experiments involving fluorescence analysis of fibronectin-coated arrays, fibronectin was labeled and purified using an Alexa Fluor 633 protein labeling kit following the manufacturer's instructions. Briefly, 0.5 ml of 1 mg/ml fibronectin in PBS was mixed with 50 μl of 1 M bicarbonate solution. The mixture was incubated with Alexa Fluor 633 reactive dye for 1 h at room temperature. The free dye was then removed from labeled fibronectin by passage through a size-exclusion column (BioRad BioGel P-30 resin, molecular weight cut-off of 40,000 D) and the number of fluorophores/fibronectin measured as described in the manufacturer's protocol (33).
To coat pallet arrays with Alexa Fluor 633-labeled fibronectin, Alexa Fluor 633-fibronectin (50 μg/ml in PBS, 1 mL) was added to control and roughened SU8 and 1002F arrays and incubated overnight (16 h) at room temperature. The coating solution was replaced with cell media after washing the arrays three times with PBS. The arrays were overlaid with DMEM with 10% fetal calf serum and incubated at 37°C for 20 days. The fluorescence of the Alexa Fluor 633-fibronectin on the pallets was measured by fluorescence microscopy (Nikon TE 300) using a cooled CCD camera (Photometrix Coolsnap FX, Tucson, AZ). Fluorescence images were collected using Metafluor software (Universal Imaging Corporation, Downingtown, PA). ImageJ (National Institutes of Health, Bethesda, MD) was used for quantitative analysis of the fluorescence micrographs. The data was normalized by subtracting the background fluorescence of uncoated SU8 or 1002F pallets.
Cell Culture
Rat basophilic leukemia (RBL), HeLa and 3T3 cells were cultured in DMEM supplemented with 10% FBS, 584 mg/L L-glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin at 37°C in a humidified 5% CO2 atmosphere. Prior to culturing cells on the micropallet arrays, the arrays with PDMS O-rings attached were sterilized by rinsing with 95% ethanol.
Micropallet Array Cell Capture Efficiency
RBL, HeLa or 3T3 cells (45,000 cells, 1.5 mL) were plated on the arrays and allowed to attach to the pallets overnight under standard tissue culture conditions. The micropallet arrays with cells were washed to remove weakly attached cells then imaged using brightfield microscopy (Nikon TE 300). The capture efficiency of different arrays for each cell line was evaluated by counting cells on the array elements as described in the results and discussion. A Student's t-test (α = 0.05) was used to determine the 95% confidence intervals of the data (to evaluate whether the differences were statistically significant).
Release and Collection of Micropallet-Containing Cells
Micropallets containing attached cells were viewed by brightfield microscopy and the pallets released using a pulsed laser as described previously (31). Briefly, a frequency-doubled Q-switched Nd:YAG laser (Minilite™ II, Continuum, Santa Clara, CA) was used to generate a single laser pulse (5-ns pulse width, 532 nm, 3 μJ and 6 μJ release energy used for SU8 and 1002F micropallets, respectively), which was spatially expanded to a 4-mm diameter (Beam Expander, Newport Corp., Irvine, CA) prior to entry into the rear port of a microscope (Nikon TE 300). An objective (Nikon Planfluor, 20x, 0.50 N.A.) was used to focus the laser pulse at the interface between the glass surface and micropallet.
Before laser-based pallet release, the array was rinsed with fresh culture medium five times to remove detached and/or weakly attached cells. The array was overlaid with fresh culture medium and a collection chamber sealed to the top surface of the array as described previously (31). This assembly was placed on the microscope stage and selected micropallet/cells were released with the pulsed laser. The assembly was then inverted to transfer the media containing released micropallet/cells into a collection chamber. The released micropallets were then inspected to determine whether the cells remained attached to the pallet surface.
Fabrication of PDMS Stamps and Microcontact Printing of SU8 and 1002F Films
A 1002F mold (100 μm square-wells, 55 μm spacing, and 50 μm depth) for forming the PDMS stamps was fabricated and coated by vapor-phase deposition with heptadecafluoro-1,1,2,2-tetrahydrodecyl trichlorosilane. The 1002F mold was cleaned with distilled water then ethanol and dried with N2. PDMS pre-polymer mixture was then layered over the 1002F mold, degassed for 1 h, and cured for 1 h at 65°C. After slowly cooling down to room temperature, the PDMS stamp was carefully cut and peeled from the 1002F master. When indicated the PDMS stamp was roughened for 25 s. After gently washing with water, then ethanol, and drying with N2, the PDMS stamps were placed in an air-plasma cleaner (Harrick PDC-001, Ithaca, New York) for 20 min immediately before use.
SU8 and 1002F films were fabricated on glass slides in a manner previously described (29). The SU8 and 1002F surfaces were roughened for 15-30 s as indicated. The films were washed with water and ethanol and then dried in a stream of N2. The photoresist surfaces were placed in an air-plasma cleaner (Harrick PDC-001) for 20 min before stamping.
A solution of 200 μg/mL BSA-Alexa Fluor 647 in PBS was spread (1 mL) on TechniCloth clean room wipes which are made of a polyester/cellulose blend (~650 mm2 area, ITW Texwipe, Mahwah, NJ) inside a culture dish and used as a stamping pad. The inking of the PDMS stamp was performed by pressing the PDMS stamp and stamping pad in contact for 10 s with the force provided by a 52 g mass. The inked stamp was depressed onto an SU8 or 1002F film for 5 s again with the force provided by a 52 g mass. The fluorescence of BSA-Alexa Fluor 647-stamped surfaces was quantified by fluorescence microscopy. The data were corrected for the background fluorescence of native SU8 and 1002F films.
RESULTS AND DISCUSSION
In-Situ Roughening of SU8 and 1002F Micropallet Arrays
Since cell and biomolecule adhesion is enhanced when surfaces are rough (34-40), we developed a simple technique for rapid in-situ roughening of microstructures such as the micropallet arrays. A key feature of the microstructures on these arrays is that the micropallets are loosely attached to their glass substrate so that laser-based lift off of desired structures is easily accomplished. Thus, the roughening process must not detach the microstructures from the array. A custom-fabricated device utilized an aqueous slurry of alumina particles to roughen the microstructures (Figure 1). The device was fabricated using a standard polystyrene dish. PDMS was used to seal a ceramic magnet to the top portion of the device which was fabricated in the base of a polystyrene dish. Arrays with the microstructures to be roughened were placed in the bottom portion of the device or lid of the polystyrene dish. A slurry of alumina particles was overlaid onto the microstructure array which was then covered with the top portion of the device. The array in the assembled device was placed on a magnetic stir plate which was used to rotate the top half of the device relative to the bottom piece with the microstructured array.
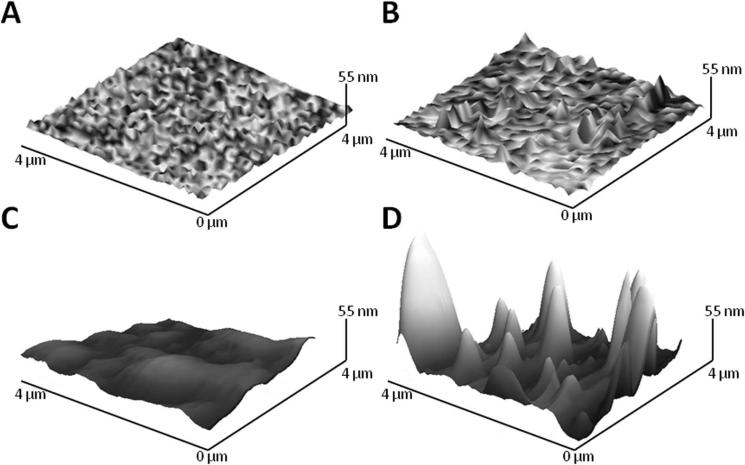
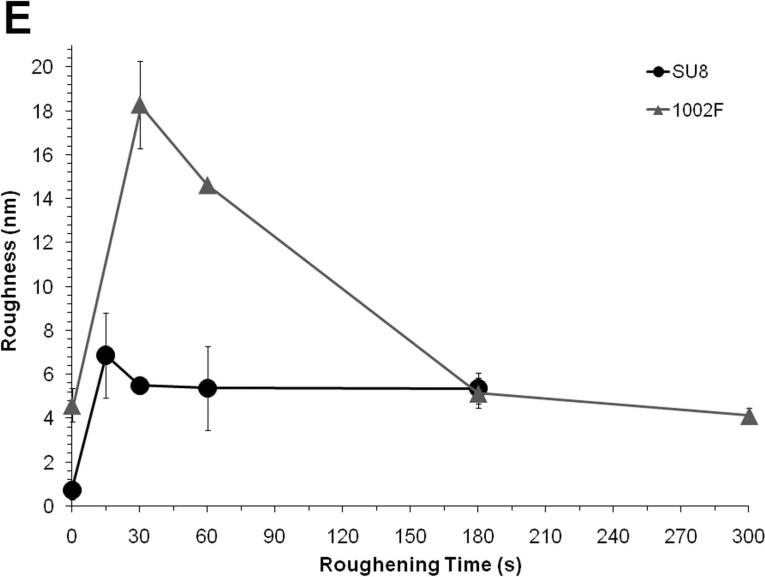
To determine whether the alumina slurry could be used to abrade the microstuctured surfaces, the micropallet arrays were polished in the presence of the alumina particles (1.0 or 0.05 μm) for varying times. When viewed by light microscopy, the surfaces roughened with 1.0-μm particles appeared free of debris (Figure S2). In contrast when surfaces were roughened with 0.05-μm particles, substantial residue remained on the array surfaces. For this reason, only 1.0-μm particles were used for subsequent experiments. The roughness of micropallet top surfaces was then measured by AFM. Pallets fabricated from SU8 or 1002F photoresist experienced a maximal increase in roughness of 9 and 4-fold, respectively after 15 to 30 s of abrasion with the alumina particles (Figure 2E). With continued polishing, the roughness declined for both the SU8 and 1002F surfaces. The AFM images were also used to view the topography of SU8 and 1002F pallets roughened for 15 and 30 s, respectively. Cone-shaped structures of nanometer size were visible on the AFM images of the roughened SU8 and 1002F micropallets (Figure 2, A-D). In contrast, native pallets did not display these distinct surface features. While SU8 and 1002F are both epoxide-based photoresists (Figure S3) with related monomer structures and identical photocatalyst, the resists possess a number of distinct properties. SU8 with 8 epoxides/monomer is an extensively cross-linked polymer whereas 1002F with only 2 epoxides/monomer is a linear uncross-linked polymer (29, 41). Consequently SU8 is a harder, less flexible material compared to 1002F. These distinct properties likely resulted in the different topography and degree of roughening of SU8 relative to 1002F surfaces.
FIGURE 2.
Shown are AFM images of native SU8 pallets (A), SU8 pallets roughened for 15 s (B), native 1002F pallets (C), and 1002F pallets roughened for 30 s (D). The variation of roughness vs. roughening time for SU8 and 1002F micropallet arrays (E) is displayed. The data points represent the average of the RMS roughness values (n = 5). The error bars represent the standard deviations of these data points.
To evaluate the uniformity of the in-situ roughening method, the roughness values for native and 15 s-roughened SU8 were measured (n=15) at the center and outer edges of a 1 cm-diameter area. Compared to native SU8 (roughness = 0.89 ± 0.18 nm), roughened SU8 showed an enhanced roughness value of 6.76 ± 1.07 nm in the center and 6.41 ± 1.11 nm around the edges the 1 cm-diameter area. These data suggest that the method resulted in uniform roughening across the photoresist surface.
The roughening of the microstructured surfaces was due to random collisions with the alumina particles in the moving fluid. It was possible that the moving fluid and particles might remove the microstructures from the underlying glass surface since no adhesive metal layer was present between the glass and photoresist. To determine whether dislodged or fragmented microstructures were present, the arrays were inspected by light microscopy. For 1002F, there was no significant increase in dislodged or fragmented micropallets on arrays roughened as long as 5 min relative to arrays that were never roughened (n >5,000 micropallets, 2 arrays) (Figure S4). For SU8 micropallet arrays, 0.10% ± 0.06 of the micropallets were dislodged or fragmented on arrays roughened for 15 s (n >5,000 micropallets, 2 arrays each photoresist) (Figure S4). The SU8 micropallet removal or damage rates increased for longer roughening times but always remained below 0.4% demonstrating the suitability of this method for in-situ roughening of microstructures. It was likely that the SU8 arrays sustained more damage than the 1002F arrays because SU8 is a harder, more brittle polymer than 1002F. The greater number of SU8 pallets removed from the glass surface compared to 1002F pallets was also consistent with the weaker adhesive force between glass and SU8 relative to glass and 1002F (29).
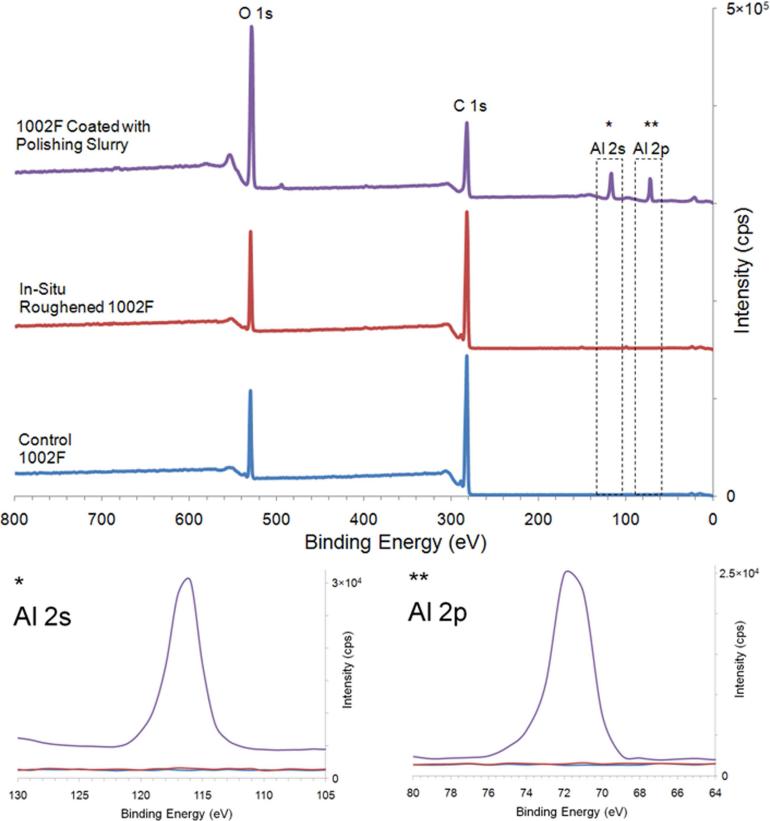
In addition to the increased surface area created by the roughening process, it is also possible that the surfaces were chemically altered. To assess this possibility, the chemical composition of the surface was analyzed using XPS. The composition of the 1002F and SU8 surfaces as judged by the peaks corresponding to C1s, O1s, and F1s was not altered by the roughening process (Figures 3, S5 and S6). However, the strength of the N1s signal from the in-situ roughened SU8 was increased relative to that of unmodified surfaces. Since nitrogen is not present in the photoresist or slurry particles, the source of the small increase in nitrogen was uncertain but appeared in all three analyzed samples. Peaks representative of Al2s and Al2p were only observed in samples for which the alumina particle slurry was intentionally left on the surfaces (Figures 3 and S6). At the binding energies indicative of Al2s and Al2, the spectra of in-situ roughened 1002F and SU8 were identical to that of native 1002F or SU8 suggesting that alumina particles did not contaminate the surface of the in-situ roughened 1002F and SU8. These data suggest that the surface chemical properties of in-situ roughened and native 1002F and SU8 were similar.
FIGURE 3.
XPS scans of an unmodified 1002F surface, an in-situ roughened 1002F surface and a 1002F surface overlaid with the particle slurry. For the lower two traces (Al 2s and 2p) the upper line is for the slurry-coated 1002F while the unmodified and in-situ roughened 1002F yielded identical traces (lower two lines).
To further characterize the surfaces, the contact angle of a water droplet on native and roughened 1002F and SU8 was measured. Native 1002F possessed a lower water-droplet contact angle compared to that for SU8 (70 ± 4 vs. 77 ± 3, n >15) consistent with the greater hydrophilicity of 1002F realtive to SU8. Roughening the 1002F and SU8 surfaces further decreased the water-droplet contact angle to 63 ± 4 and 66 ± 4, respectively. The decrease in contact angle following roughening is consistent with that observed for other hydrophilic materials (42-44).
Cell Adhesion to Roughened SU8 and 1002F Micropallet Arrays
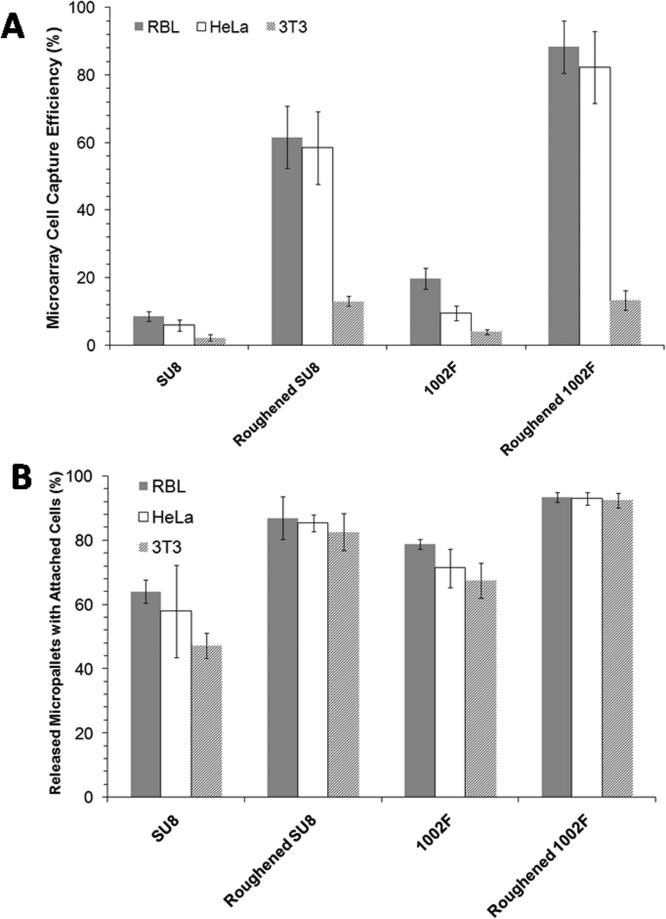
Most cell types must attach to a surface to grow and divide (45). Surface topography is well known to influence cell adhesion with rougher surfaces (in an appropriate range) enhancing attachment (38). To determine whether the microstructures roughened with the alumina particles offered improved cell adhesion, three cell lines (RBL, HeLa, and 3T3 cells) were cultured on SU8 and 1002F micropallet arrays. These three cells lines were selected due to their different tissue origin and phenotype. Cells were plated on native and roughened array surfaces at identical cell densities, cultured for 16 h, and then examined by microscopy (Figure S7). The capture efficiency for the cells on the arrays was defined as the number of cells captured on the micropallets divided by the total number of cells added to the array chamber. For roughened SU8 arrays, a 6-10x increase in cell capture efficiency was observed relative to native arrays for all three cell types (p <10-7) (Figure 4A). For 1002F arrays, the improvement in capture efficiency for roughened vs. native arrays was 3-9 fold for the three different cell lines (p <0.0005) (Figure 4A). These results are in agreement with previous reports demonstrating that rougher surfaces increased cell capture and growth rate (46-52).
FIGURE 4.
(A) The cell capture efficiency of native and roughened arrays composed of SU8 or 1002F. The data points represent the average value (n > 200 micropallets) and the error bars represent the 95% confidence intervals as determined by a Student's t-test (α = 0.05). At least ten random micrographs each one containing 20 micropallets was inspected per array (n ≥ 2 arrays). (B) Attachment of HeLa, 3T3, and RBL cells to micropallets following laser-based pallet release for unroughened and roughened SU8 and 1002F arrays. (n ≥ 25 micropallets, at least two arrays per condition).
As demonstrated in previous work (53), a major advantage of the micropallet arrays is their use for cell separations for biomedical research. For effective cell sorting, the cells must remain attached to the micropallet during its laser-based release from the underlying glass substrate. To study the effect of roughening on cell attachment during laser release, the percentage of cells remaining attached to roughened and native pallets after pallet release was measured. The 3T3, HeLa, and RBL cells were cultured on the arrays of SU8 or 1002F micropallets overnight. A single laser pulse (532 nm, 5 ns) was used to release pallets with attached cells from the array. The released micropallets were observed immediately for the presence or absence of an attached cell. The percent of released pallets with an attached cell was recorded. For all three cell types, the roughened surfaces were more efficient at retaining the adherent cells compared to the native photoresist. On average 50% and 30% more cells remained on the roughened SU8 and 1002F, respectively, relative to their non-roughened counterparts after release (Figure 4B). These differences were significant with p ≤ 0.05. The greater degree of improvement for SU8 relative to 1002F is likely due to the fact that the surface of native SU8 is smoother on average than that of 1002F and therefore roughening has a greater effect. Of the three cell types tested, the 3T3 cells benefited the most from adhesion to roughened surfaces during laser release. 3T3 cells demonstrate a loose adhesion to many culture surfaces, for example, polystyrene, whereas HeLa and RBL cells are tightly adherent to standard culture surfaces. Thus it is perhaps not surprising that roughened surfaces improved the collection of 3T3 cells more than that of RBL or HeLa cells.
Coating Stability of Roughened SU8 and 1002F Micropallet Arrays
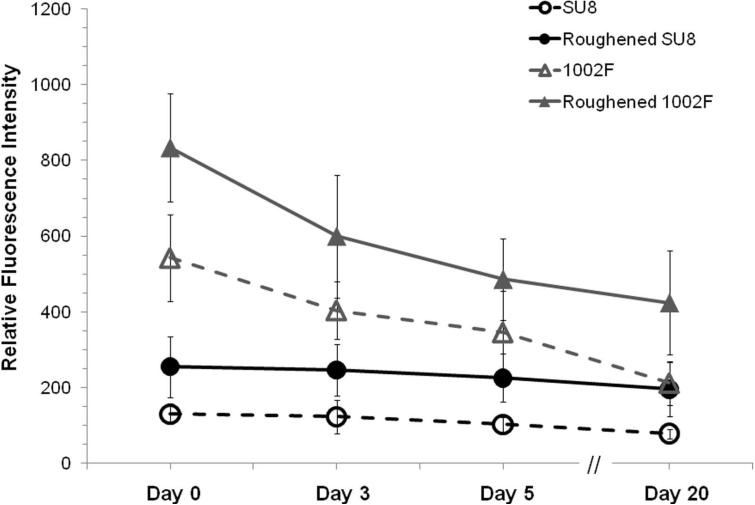
In most cases, micropallet arrays must be coated with gels or proteins such as fibronectin prior to culture of cells on the arrays (29-31, 53-57). Many experiments require that the cells grow on the arrays for several days to weeks during which time the coated layer must remain stably attached to the pallet surface. This is especially critical for primary cells, such as stem cells, where the lack of a coating layer can result in cell death or differentiation (55). To determine whether surface roughening of the arrays might increase the density of these base coatings, Alexa Fluor 633-labeled fibronectin was incubated with native and roughened SU8 and 1002F micropallet arrays. The arrays were placed in tissue culture medium and the fluorescence intensity of the micropallets was measured over time by microscopy (Figure 5). At day 0 of incubation in the aqueous medium, the normalized fluorescence intensity of the roughened SU8 and 1002F arrays was more than 2 and 1.5 times, respectively, relative to that for native arrays. 1002F arrays possessed significantly more surface fibronectin than SU8. This may be due to the increased surface roughness relative to that of SU8 or the presence of hydroxyl groups in 1002F not present in SU8 (Figure S3) (29, 58). By day 20, the fluorescence of all of the arrays had decreased relative to that at day 0. However the roughened arrays possessed fluorescence intensities 2-2.5 times that of the native arrays. The rate of Alexa Fluor 633-labeled fibronectin loss between day 0 and 20 was similar for native arrays and roughened arrays suggesting that the larger quantity of fibronectin on the roughened arrays was due to the greater surface area relative to that of the unroughened arrays. These results were also consistent with the XPS data which suggested that the roughened and unroughened surfaces possessed similar chemical properties.
FIGURE 5.
The efficiency and stability over time for Alexa Fluor 633-labeled fibronectin on native and roughened SU8 and 1002F micropallet arrays (n ≥ 32 micropallets from two arrays) is shown. The average of the data is shown with the error bars representing the standard deviations of the data points.
Improved Efficiency of μCP Transfer of Coatings onto Roughened Surfaces
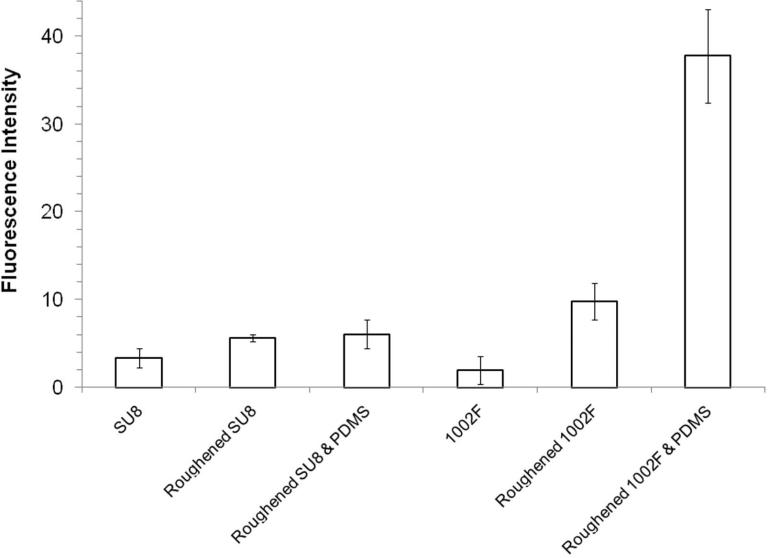
Despite the advantages that μCP provides (59-62), especially for polymeric substrates, the surfaces must be well-prepared for efficient and homogeneous transfer of the patterns from the stamp, a task which is difficult to achieve. The plasma treatment of both the PDMS stamp and the polymeric substrate before stamping facilitates ink transfer (63), but a simple solution that further enhances the efficiency and homogeneity of stamping is highly desirable. Consequently, the influence of roughening on both the stamp and the receiving substrate on ink transfer by μCP was measured. A PDMS stamp was used to transfer BSA-Alexa Fluor 647 to SU8 or 1002F substrates. The transfer of fluorescent BSA was measured for roughened and native PDMS stamps as well as roughened and native SU8 and 1002F substrates. The fluorescence of the stamped BSA-Alexa Fluor 647 on SU8 and 1002F substrates was measured by fluorescence microscopy (Figures 6 and S8). Roughening only the SU8 and 1002F films increased the amount of printed BSA-Alexa Fluor 647 by 2 and 5-fold, respectively, relative to native SU8 and 1002F. Thus mechanically roughened surfaces can demonstrate increased acceptance of ink from the stamp. Roughening both the PDMS stamp and the SU8 substrate further enhanced transfer of the BSA-Alexa Fluor 647 by 8% relative to roughened SU8 alone. The efficiency of BSA-Alexa Fluor 647 transfer from a roughened PDMS stamp to a roughened 1002F surface was improved by 20-fold compared to unroughened surfaces. This dramatic enhancement was likely due to both the increased surface area of the hydrophilic 1002F and hydrophilic oxidized PDMS.
FIGURE 6.
Fluorescence intensity of BSA-Alexa Fluor 647 printed on SU8 or 1002F films before and after roughening of the substrate or stamp. The average fluorescence intensity of the printed squares (100 × 100 μm, n ≥ 240) is shown and the error bars represent the standard deviations of the data points.
CONCLUSION
Mechanical roughening of polymeric microstructures with a particle slurry was reported. A simple device using a magnetic stir bar yielded controlled roughening of microstructure surfaces. Aqueous slurries of particles in a variety of sizes and of varying composition are commercially available at a low cost. Thus the roughening process can be tailored to increase or decrease the roughness of miniaturized polymeric structures and surfaces to the desired degree by optimizing the polishing time as well as the particle properties. This in-situ roughening technique was successfully used in two applications for polymeric microstructures: i) improving biomolecule and cell adhesion to micropallets utilized for cell sorting, and ii) enhancing the efficiency of μCP to pattern biomolecules on surfaces. In addition to these applications, this mechanical polishing method should find widespread utility for other fragile structures such as cantilevers and for the placement of biomolecule or cell coatings on surfaces for a variety of biomedical applications (2, 64-67).
Supplementary Material
Acknowledgement
This research was supported by the National Institutes of Health (EB007612). We greatly acknowledge the technical support provided by Professors Mark Schoenfisch and Dorothy Erie (UNC Department of Chemistry), Dr. Carrie Donley (Chapel Hill Analytical and Nanofabrication Laboratory) and also the UNC-Michael Hooker Microscopy Facility (Dr. Michael Chua) regarding AFM and XPS analyses. Jocelyn Wang and Annadele Herman are acknowledged for their laboratory assistance.
Footnotes
Supporting Information Available: Additional figures are included: a photograph of the roughening apparatus, DIC micrographs representing micro-structure and surface damage and/or contamination during roughening, a graph of micropallet removal vs. the roughening time, XPS scans for 1002F and SU8 surfaces, micrographs of cell growth on control and roughened arrays, and fluorescence micrographs of stamped features on 1002F and SU8 surfaces. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Sato K, Mawatari K, Kitamori T. Lab. Chip. 2008;8:1992–1998. doi: 10.1039/b814098g. [DOI] [PubMed] [Google Scholar]
- 2.Wang W, Soper SA. Bio-MEMS: Technologies and Applications. CRC Press; Boca Raton: 2007. p. 477. [Google Scholar]
- 3.Soper SA, Ford SM, Qi S, McCarley RL, Kelly K, Murphy MC. Anal. Chem. 2000;72:643A–651A. doi: 10.1021/ac0029511. [DOI] [PubMed] [Google Scholar]
- 4.Shadpour H, Musyimi H, Chen J, Soper SA. J. Chromatogr. A. 2006;1111:238–251. doi: 10.1016/j.chroma.2005.08.083. [DOI] [PubMed] [Google Scholar]
- 5.Fan ZH, Ricco AJ. BioMEMS Biomed. Nanotech. 2006;2:311–328. [Google Scholar]
- 6.West J, Becker M, Tombrink S, Manz A. Anal. Chem. 2008;80:4403–4419. doi: 10.1021/ac800680j. [DOI] [PubMed] [Google Scholar]
- 7.Yi C, Li C-W, Ji S, Yang M. Anal. Chim. Acta. 2006;560:1–23. [Google Scholar]
- 8.Erickson D, Li D. Anal. Chim. Acta. 2004;507:11–26. [Google Scholar]
- 9.Huang C, Le Y.-c. TW Patent 2002-91124050-551008 Circuit scraping rework method and the slurry thereof. 2003
- 10.Ahn Y, Yoon J-Y, Baek C-W, Kim Y-K. Wear. 2004;257:785–789. [Google Scholar]
- 11.Kourouklis C, Kohlmeier T, Gatzen HH. Sens. Actuators A. 2003;106:263–266. [Google Scholar]
- 12.Lee S-D, Nien C-F. US Patent 2002-76395-6548409 Method of reducing micro-scratches during tungsten CMP. 2003
- 13.Chen H.-h., Wei Y-T, Yang M-S. US Patent 2000-483581-6380069 Polishing with chemical-mechanical slurry stages for removal of micro-scratch markings on a metal layer for electric integrated circuits. 2002
- 14.Lee C-T, Ferguson KA, Herreria EV. EP Patent 99-309936-1008571 Methods of treating ceramic surfaces. 2000
- 15.Wang ZL, Gao P, Ding Y. WO Patent 2006-US34758-2007081409 Super-elastic helical nanostructures and methods of fabrication and use. 2007
- 16.Teh WH, Luo JK, Liang CT, Smith CG. MEMS/NEMS. 2006;2:173–221. [Google Scholar]
- 17.Bower RW. US Patent 2006-335162-2006264014 Structures, materials and methods for fabrication of nanostructures. 2006
- 18.Chua SJ, Chen P, Wang Y. WO Patent 2004-SG274-2006025793 Nanostructures and method of making the same. 2006
- 19.Ramamoorthi S, Mardilovich P, Kornilovich P, Korthuis VC. US Patent 2005-215985-2006003267 Nanostructure and method of fabricating nanostructures. 2006
- 20.Sarajlic E, Berenschot JW. WO Patent 2004-NL902-2005061377 Method for manufacturing a nanostructure. 2005
- 21.Mijatovic D, Eijkel JCT, Van den Berg A. Lab. Chip. 2005;5:492–500. doi: 10.1039/b416951d. [DOI] [PubMed] [Google Scholar]
- 22.Carpenter SE. EP Patent 2003-29781-1491493 Nanostructure fabrication using microbial mandrel. 2004
- 23.Helt JM. WO Patent 2004-US10932-2004092836 Fabrication of nanostructures and nanowires. 2004
- 24.Imada A, Den T. WO Patent 2004-JP1281-2004070709 Structure, optical device, magnetic device, magnetic recording medium and manufacturing method thereof. 2004
- 25.Kim S.-g., El Aguizy TA, Jeong J.-h., Jeon Y. WO Patent 2003-US32109-2004033370 Nanopellets and method of making nanopellets for nanostructure fabrication. 2004
- 26.Schaper CD. US Patent 2002-246379-2003219992 Replication and transfer of microstructures and nanostructures. 2003
- 27.Hyldgaard P, Chakarov D, Lundqvist B. WO Patent 2002-SE2461-2003055793 A method for manufacturing a nanostructure in-situ, and in-situ manufactured nanostructure devices. 2003
- 28.Matsui S. In: Handbook of Nanostructured Materials and Nanotechnology. Nalwa H, editor. Academic Press; London: 2001. p. 834. [Google Scholar]
- 29.Pai J-H, Wang Y, Salazar GTA, Sims CE, Bachman M, Li GP, Allbritton NL. Anal. Chem. 2007;79:8774–8780. doi: 10.1021/ac071528q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Sims CE, Marc P, Bachman M, Li GP, Allbritton NL. Langmuir. 2006;22:8257–8262. doi: 10.1021/la061602k. [DOI] [PubMed] [Google Scholar]
- 31.Salazar G. T. a., Wang Y, Young G, Bachman M, Sims CE, Li GP, Allbritton NL. Anal. Chem. 2007;79:682–687. doi: 10.1021/ac0615706. [DOI] [PubMed] [Google Scholar]
- 32.Whitehouse DJ. Handbook of Surface Metrology. Institute of Physics Publishing; Bristol: 1994. p. 350. [Google Scholar]
- 33.Invitrogen Alexa Fluor® 633 Protein Labeling Kit. 2004 http://probes.invitrogen.com/media/pis/mp20170.pdf.
- 34.Anderson DC, Springer TA. Annu. Rev. Med. 1987;38:175–94. doi: 10.1146/annurev.me.38.020187.001135. [DOI] [PubMed] [Google Scholar]
- 35.Buck CA, Horwitz AF. Annu. Rev. Cell Biol. 1987;3:179–205. doi: 10.1146/annurev.cb.03.110187.001143. [DOI] [PubMed] [Google Scholar]
- 36.Curtis A, Wilkinson C. Biomaterials. 1998;18:1573–1583. doi: 10.1016/s0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 37.Frey MT, Tsai IY, Russell TP, Hanks SK, Wang Y.-l. Biophys. J. 2006;90:3774–3782. doi: 10.1529/biophysj.105.074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris AK., Jr. J. Biomech. Eng. 1984;106:19–24. doi: 10.1115/1.3138449. [DOI] [PubMed] [Google Scholar]
- 39.Hynes RO. Cell. 1987;48:549–54. doi: 10.1016/0092-8674(87)90233-9. [DOI] [PubMed] [Google Scholar]
- 40.Mustafa K, Oden A, Wennerberg A, Hultenby K, Arvidson K. Biomaterials. 2005;26:373–381. doi: 10.1016/j.biomaterials.2004.02.037. [DOI] [PubMed] [Google Scholar]
- 41.Shaw JM, Gelorme JD, LaBianca NC, Conley WE, Holmes SJ. IBM J. Res. Dev. 1997;41:81–94. [Google Scholar]
- 42.Chow TS. J. Phys.: Cond. Matt. 1998;10:L445–L451. [Google Scholar]
- 43.Hao L, Lawrence J, Phua YF, Chian KS, Lim GC, Zheng HY. J. Biomed. Mater. Res. B. 2005;73:148–156. doi: 10.1002/jbm.b.30194. [DOI] [PubMed] [Google Scholar]
- 44.Hong KT, Imadojemu H, Webb RL. Exp. Therm. Fluid Sci. 1994;8:279–285. [Google Scholar]
- 45.Shapiro HM. Practical Flow Cytometry. Wiley & Sons, Inc.; Hoboken: 2003. p. 736. [Google Scholar]
- 46.Widdicombe JH, Sachs LA, Finkbeiner WE. Vitro Cell Dev. Biol. Anim. 2003;39:51–5. doi: 10.1290/1543-706X(2003)039<0051:EOGSOD>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 47.Kumakura M, Kaetsu I. J. Mater. Sci. Lett. 1985;4:761–3. [Google Scholar]
- 48.Ma T, Li Y, Yang S-T, Kniss DA. Biotechnol. Progr. 1999;15:715–724. doi: 10.1021/bp990072y. [DOI] [PubMed] [Google Scholar]
- 49.Weis RP, Montchamp J-L, Coffer JL, Attiah DG, Desai TA. Dis. Markers. 2002;18:159–165. doi: 10.1155/2002/727014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krasteva N, Seifert B, Albrecht W, Weigel T, Schossig M, Altankov G, Groth T. Biomaterials. 2004;25:2467–2476. doi: 10.1016/j.biomaterials.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 51.Kim H-K, Jang J-W, Lee C-H. J. Mater. Sci. - Mater. Med. 2004;15:825–830. doi: 10.1023/b:jmsm.0000032824.62866.a1. [DOI] [PubMed] [Google Scholar]
- 52.Johansson F, Kanje M, Eriksson C, Wallman L. Phys. Status Solidi C. 2005;2:3258–3262. [Google Scholar]
- 53.Wang Y, Young G, Aoto Phillip C, Pai J-H, Bachman M, Li GP, Sims Christopher E, Allbritton Nancy L. Cytometry A. 2007;71:866–74. doi: 10.1002/cyto.a.20424. [DOI] [PubMed] [Google Scholar]
- 54.Shadpour H, Sims CA, Allbritton NL. Cytometry A. 2009;75:609–618. doi: 10.1002/cyto.a.20741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shadpour H, Sims CA, Thresher RJ, Allbritton NL. Cytometry A. 2009;75:121–129. doi: 10.1002/cyto.a.20672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Salazar G. T. a., Pai J-H, Shadpour H, Sims CE, Allbritton NL. Lab. Chip. 2008;8:734–740. doi: 10.1039/b800286j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Young G, Bachman M, Sims CE, Li GP, Allbritton NL. Anal. Chem. 2007;79:2359–2366. doi: 10.1021/ac062180m. [DOI] [PubMed] [Google Scholar]
- 58.Keselowsky BG, Collard DM, Garcia AJ. J. Biomed. Mater. Res. A. 2003;66A:247–259. doi: 10.1002/jbm.a.10537. [DOI] [PubMed] [Google Scholar]
- 59.Perl A, Reinhoudt DN, Huskens J. Adv. Mater. 2009;21:2257–2268. [Google Scholar]
- 60.Ruiz SA, Chen CS. Soft Matter. 2007;3:168–177. doi: 10.1039/b613349e. [DOI] [PubMed] [Google Scholar]
- 61.Menard E, Rogers JA. In: Springer Handbook of Nanotechnology. Bhushan B, editor. Springer; New York: 2007. p. 1916. [Google Scholar]
- 62.Quist AP, Pavlovic E, Oscarsson S. Anal. Bioanal.Chem. 2005;381:591–600. doi: 10.1007/s00216-004-2847-z. [DOI] [PubMed] [Google Scholar]
- 63.Schmalenberg KE, Buettner HM, Uhrich KE. Biomaterials. 2004;25:1851–1857. doi: 10.1016/j.biomaterials.2003.08.048. [DOI] [PubMed] [Google Scholar]
- 64.Zougagh M, Rios A. Analyst. 2009;134:1274–1290. doi: 10.1039/b901498p. [DOI] [PubMed] [Google Scholar]
- 65.Alvarez M, Zinoviev K, Moreno M, Lechuga LM. Opt. Biosens. 2008:419–452. [Google Scholar]
- 66.Nordstrom M, Keller S, Lillemose M, Johansson A, Dohn S, Haefliger D, Blagoi G, Havsteen-Jakobsen M, Boisen A. Sensors. 2008;8:1595–1612. doi: 10.3390/s8031595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Addae-Mensah KA, Wikswo JP. Exp. Biol. Med. 2008;233:792–809. doi: 10.3181/0710-MR-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.