Fig. 4.
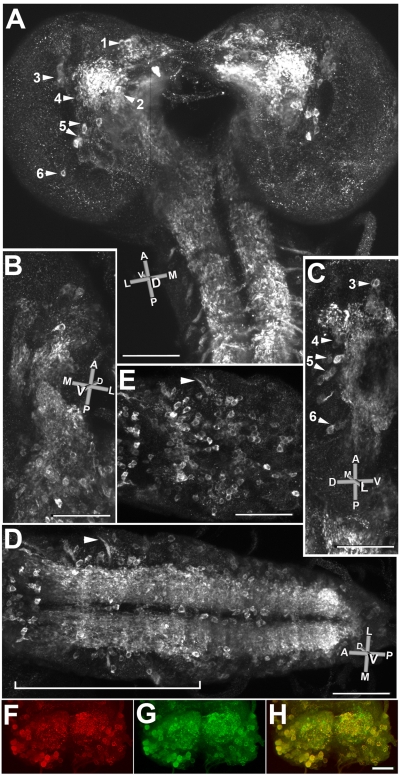
Expression of dVGAT protein in larvae. (A) Confocal stacks from three combined, ×40 fields show a dorsal view of the brain and the anterior portion of the nerve cord. An orientation key is shown: L. lateral; M, medial; D, dorsal; V, ventral; A, anterior; P, posterior. (B) A ventral view of one half of the brain and the anterior portion of the nerve cord from a separate larva. (C) A side view (lateral to medial) of another larva showing the central brain. A small portion of the anterior nerve cord is visible at the bottom of the panel. Six clusters of GABAergic neurons are visible in the brain, labeled 1–6 in A, with clusters 3–6 visible in C. Note that cluster 2 in A is obscured by the intense labeling of the processes. (D) A ventral view of the nerve cord from another animal including optical sections through the entire depth of the nerve cord. A nerve root is indicated (arrowhead). (E) A subset of optical sections from the bracketed region in D reveals labeled cells on the ventral surface of the cord, with approx. 75 cells per abdominal segment. For comparison, in late stage embryos labeled with an antiserum to GABA, each segment was estimated to contain approx. 54 strongly and 50 weakly labeled neurons, ~20% of the total number of neurons per segment (Kuppers et al., 2003). (F–H) Co-labeling for dVGAT in red (F) and GABA in green (G) shows extensive co-localization (H, yellow) in both cell bodies and neuropil. As in D and E most cell bodies are near the ventrolateral surfaces of the cord. Scale bars, (A–E) 100 μm; (F–H) 50 μm.