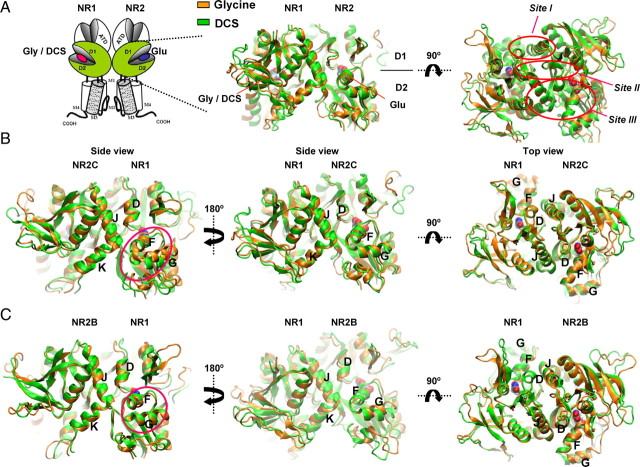
Figure 2.
Molecular dynamics simulations of the NR1/NR2B and NR1/NR2C ligand binding domain dimer. A, Schematic showing the NMDA receptor subunit arrangement with the ligand binding domains of NR1 and NR2 subunit shown in green. Homology models of NR1/NR2B and NR1/NR2C were based on the crystal structure of NR1/NR2A (Furukawa et al., 2005); molecular dynamics simulations were run on the hydrated protein with glycine (orange) or DCS (green) docked into the NR1 pocket and glutamate in the NR2 pocket. Dimer interaction sites Site I-III are indicated by red circles. B, Molecular dynamics simulations of NR1/NR2C ligand binding domain dimer. The difference in orientation of helix F and G of NR1 is evident (circled in side view). In addition, differences in arrangement of helices F and K of NR2C can be seen. C, Molecular dynamics simulations of NR1/NR2B ligand binding domain dimer. The difference in orientation of helices F and G of NR1 is evident (circled in side view).