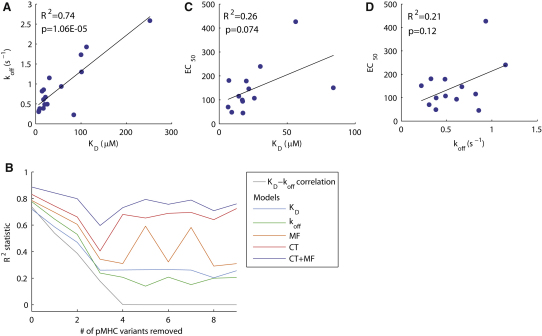
Figure 4.
Analysis of Model Fitting to Data Subsets with Larger Variability in the On Rate
(A) The 17 pMHC variants that comprise the entire data set exhibit a large correlation between KD and koff (R2 = 0.74). By systematically removing a fixed number of pMHC variants, we identified the subset that minimized the KD-koff correlation. For example, we calculated the correlation for all 17 possible subsets of 16 pMHC variants and selected the subset with the smallest KD-koff correlation.
(B) The square of the KD-koff correlation as a function of the number of pMHC variants removed for the subset with the smallest correlation. In this way, we generated data sets with lower KD-koff correlations and hence larger variability in the on rate. Also shown in (B) is the R2 statistic for fits of the KD, koff, molecular flexibility (MF), confinement time (CT), and the combined model (MF+CT) to all subsets considered. Computations for subsets consisting of less than eight pMHC variants did not provide reliable results because the number of parameters approached the number of data points.
(C and D) The correlations of pMHC potency with KD and koff for the subset of 13 pMHC variants (i.e., when 4 pMHC variants are removed). See Figure 5 for correlations with other models. In Table 2 we report the p values associated with each fit, showing that when removing more than two pMHC variants, correlations with KD and koff are not significant (p > 0.05). Also shown in Table 2 are p values associated with an F-test that determines whether the additional parameter(s) in the CT, MF, and combined model is significant. The fitted parameters from each model are listed in Table S2.