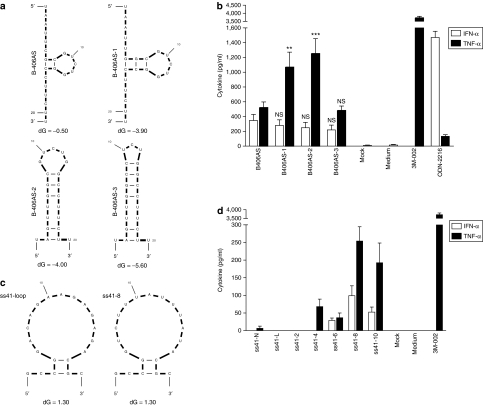
Figure 1.
Relative immunostimulation of ssRNAs with sequence or structural alterations. (a,b) mFOLD-predicted structure of ssRNAs at 37 °C (ref. 44). All ss41-RNAs have a similar predicted secondary structure, and only ss41-loop and ss41-8 are presented here. The amount of uridines in the ss41 series is indicated by the x in the ss41-x name. Predicted free-energy of the loops is indicated with dG value. ss41-Native does not have a predicted secondary structure. All sequences are shown in Table 1. (c,d) Immunostimulation of ssRNAs in human peripheral blood mononuclear cells treated with 90 nmol/l of indicated ssRNA complexed with N-[1-(2,3-dioleoyloxy)propyl]-N,N, Ntrimethylammonium methylsulfate and incubated for 16 hours at 37 °C. Cytokine production was measured by specific enzyme-linked immunosorbent assay as described in Materials and Methods. The data are averaged from (c) three independent experiments in biological triplicate from four blood donors and (d) two independent experiments in biological triplicate from three blood donors. (c) Unpaired two-tailed t-test and nonparametric two-tailed Mann–Whitney test were used for IFN-α and TNF-α statistical comparison compared with B-406AS, respectively. IFN, interferon; NS, nonsignificant; ssRNA, single-stranded RNA; TNF, tumor necrosis factor.