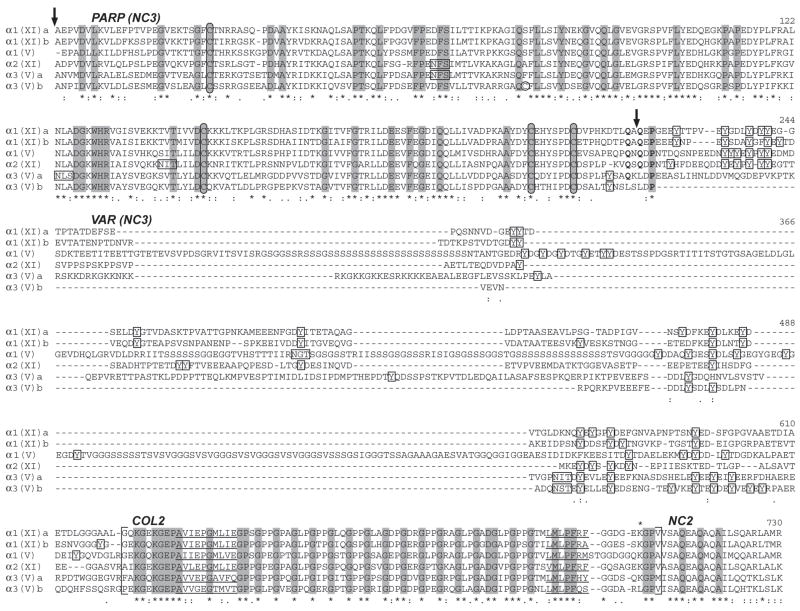
Fig. 2.
Alignment of NH2-terminal sequences of zebrafish clade B procollagen chains. NH2-terminal sequences were aligned using the EMBL-EBI ClustalW2 server. Dashes represent gaps introduced for optimal sequence alignment. Vertical arrows mark the approximate site of signal peptide cleavage and cleavage by BMP1-like proteinases, based on predicted and demonstrated sites in mammalian clade B chains (Gopalakrishnan et al., 2004; Greenspan et al., 1991; Imamura et al., 2000; Imamura et al., 1998; Unsold et al., 2002). PARP, and variable (VAR) subdomains of noncollagenous domain 3 (NC3) are labeled, as is collagenous domain 2 (COL2), and noncollagenous domain 2 (NC2). The extent of COL2 is marked by brackets. Noncollagenous interruptions in the COL2 domain are underlined. Cysteines are circled. Tyrosines between the PARP and COL2 domains are boxed, as are potential Asn-linked glycosylation sites. Residues found at the pro-α1(V) BMP1-cleavage site and conserved in zebrafish clade B chains are in boldface type. Asterisks and dots at the bottom of the alignment signify extent of similarity of aligned sequences, with asterisks denoting identity at a given position in all six zebrafish clade B chains. Residues identical in all six zebrafish and all reported mammalian clade B procollagen chains (Imamura et al., 2000) are shaded.