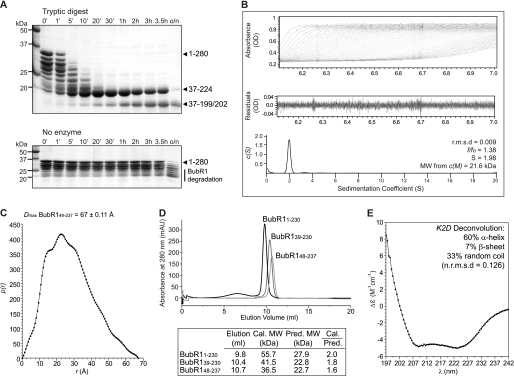
FIGURE 2.
Biochemical and biophysical characterization of the BubR1 N-terminal region. A, limited proteolysis of BubR1(1–280) identified protease-resistant fragments of ∼18 and 20 kDa. These fragments encompass residues Ile37–Arg199 or Arg202 and Ile37–Arg224, respectively. B, sedimentation velocity analysis of BubR1(48–237) reveals it to be an asymmetrical monomer. Using absorbance data (top), the sedimentation coefficient distribution c(S) (bottom) was calculated using SEDFIT to minimize residuals (middle) and with a root mean square deviation (r.m.s.d.) of less than 0.01. The resultant frictional coefficient (f/f0), sedimentation coefficient (S), and molecular weight (MW) are shown. C, small angle x-ray scattering of BubR1(48–237) measures a Dmax of 67 ± 0.11 Å, indicating an elongated shape. The intra-particle distance distribution function p(r) was calculated using GNOM. D, analytical size exclusion chromatography of BubR1 proteins shows that the calculated (cal.) to predicted (pred.) molecular weight ratio increases as more of the N-terminal of BubR1 is included. mAU, milli-absorbance units. E, far-UV circular dichroism spectra of BubR1(48–237) reveal a predominantly α-helical structure.