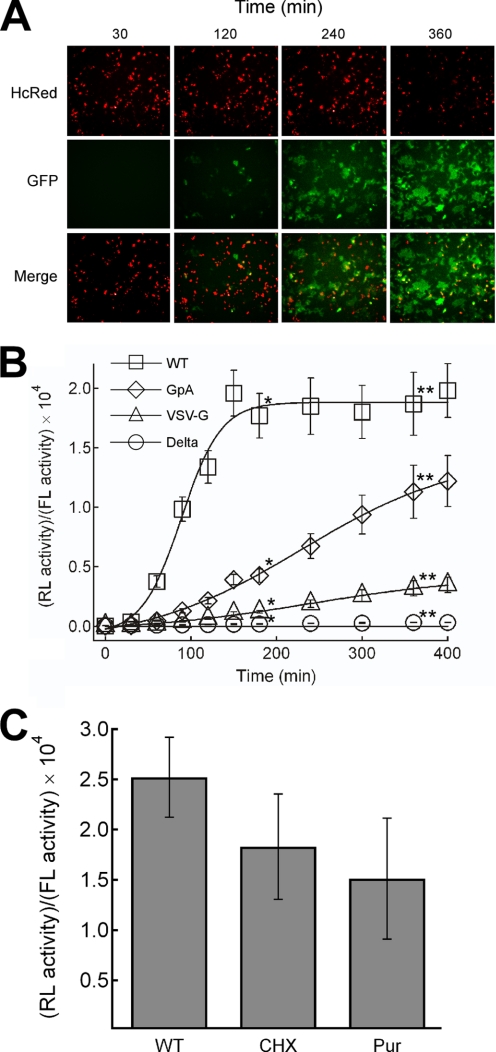
FIGURE 3.
Real-time monitoring of HIV-1 Env-mediated fusion. A, monitoring of GFP signal after co-culture of 293FT cell harboring wild type Env and DSP1–7 with 293CD4 cells harboring DSP8–11. Images were obtained with IN Cell Analyzer 1000. The Env-expressing cells bear red fluorescence in nuclei derived from the HcRed included in the Env expression vector (HcRed) (10). The views of the green signal (GFP) and merged images (Merge) are also shown. B, monitoring of the RL activity is shown. The recovered RL activity was divided by the firefly luciferase (FL) activity to normalize the transfection efficiency of the Env expression vector as described previously (15). Open squares, WT Env of HIV-1; open diamonds, GpA mutant; open triangles, the VSV-G mutant. Delta depicts the vector with the deleted envelope gene (open circle). * and **, statistically significant by independent t test (p < 0.0001). C, shown is the effect of protein synthesis inhibitor on the efficiency of fusion in DSP-mediated system. Cycloheximide (CHX, 100 μm) and puromycin (Pur, 10 μm) were added immediately before co-cultivation. The RL activities were measured 200 min after co-cultivation. The values were normalized by dividing them with the firefly luciferase activities and compared with that in the absence of the inhibitors (WT).