Abstract
Reversibly switchable fluorescent proteins can be repeatedly photoswitched between a fluorescent and a nonfluorescent state by irradiation with the light of two different wavelengths. The molecular basis of the switching process remains a controversial topic. Padron0.9 is a reversibly switchable fluorescent protein with “positive” switching characteristics, exhibiting excellent spectroscopic properties. Its chromophore is formed by the amino acids Cys-Tyr-Gly. We obtained high resolution x-ray structures of Padron0.9 in both the fluorescent and the nonfluorescent states and used the structural information for molecular dynamics simulations. We found that in Padron0.9 the chromophore undergoes a cis-trans isomerization upon photoswitching. The molecular dynamics simulations clarified the protonation states of the amino acid residues within the chromophore pocket that influence the protonation state of the chromophore. We conclude that a light driven cis-trans isomerization of the chromophore appears to be the fundamental switching mechanism in all photochromic fluorescent proteins known to date. Distinct absorption cross-sections for the switching wavelengths in the fluorescent and the nonfluorescent state are not essential for efficient photochromism in fluorescent proteins, although they may facilitate the switching process.
Keywords: Fluorescence, Microscopic Imaging, Molecular Dynamics, Phototransduction, Spectroscopy, X-ray Crystallography, Fluorescent Protein, Light-induced Switching, Photochromic Protein
Introduction
Green fluorescent proteins and GFP-like proteins (fluorescent proteins, FPs)3 have become important tools for dissecting internal processes in cells and organisms, such as the monitoring of cellular ion concentrations and pH, analyzing gene expression, tracking protein movement, the migration of pathogens within a host, and many others (1–6). Recently, derivatives of FPs have been described whose fluorescence properties may be modulated by irradiation (7, 8). Three classes of switchable FPs may be distinguished, namely i) photoactivatable FPs that can be irreversibly switched from a dark to a fluorescent state, ii) photoconvertable FPs where one fluorescent color can be changed to another, and iii) photochromic or reversibly switchable FPs (RSFPs) enabling repeated on/off switching.
RSFPs can be reversibly switched between a fluorescent (on) and a nonfluorescent (off) state by irradiation with light of two different wavelengths. One of the wavelengths concomitantly induces fluorescence in the on state. RSFPs were initially isolated from the sea anemone Anemonia sulcata (9). However, the original protein (asFP595) exhibited only marginal fluorescence and forms tetramers. Recently, improved photochromic proteins were engineered based on proteins from the corals Pectiniidae (Dronpa and variants) (10–12), and Clavularia (mTFP0.7) (13), as well as from the FP mCherry (14) (rsCherryRev). RSFPs can be grouped into those with positive switching characteristics (the wavelength that induces fluorescence switches the protein from the off to the on state) and those with negative switching characteristics (the wavelength that induces fluorescence switches the protein from the on to the off state).
Several studies address the molecular mechanism of switching in these RSFPs (11, 13, 15–21). Taken together, these data point to a cis-trans isomerization, often accompanied by a change in the protonation state of the chromophore as the structural basis for reversible switching in RSFPs. Interestingly, a recent NMR study indicates that the protonated off state of Dronpa exhibits increased flexibility, suggesting that the flexibility and the protonation state of the chromophore, rather than isomerization, are the determinants for effective switching (22). Hence, the actual switching mechanism remains a controversial topic, and currently very little is known about the protonation states of the amino acid residues surrounding the chromophore.
Here, we present the three-dimensional structure of the RSFP Padron0.9 in the fluorescent state at 1.65 Å resolution as well as in the nonfluorescent state at 1.80 Å resolution. We have focused our attention on this protein because its outstanding spectroscopic and switching properties (12) make this protein an exemplary RSFP with positive switching characteristics. Furthermore, we compare Padron0.9 with Dronpa, an example of an RSFP with negative switching characteristics (10, 11, 17). Padron0.9 and Dronpa differ only in a few amino acid residues, are structurally highly similar, but exhibit antipodal switching characteristics. Thus, the comparison of these two proteins provides a powerful base to study the molecular basis of light-driven reversible switching. Moreover, we used the crystallographic data of Padron0.9 for molecular dynamics simulations to investigate the protonation states of the amino acid residues within the chromophore pocket during the switching process. We find that a cis-trans isomerization of the chromophore is the structural basis for the switching in Padron. This study further demonstrates that different absorption characteristics of the fluorescent and the nonfluorescent chromophore, evoked by distinct protonation states, are not a general requirement for photochromic switching in RSFPs.
EXPERIMENTAL PROCEDURES
Protein Production and Crystallographic Analyses
A pQE31 (Qiagen) expression vector containing the coding sequence for Padron0.9 was transformed into the Escherichia coli strain SURE (Stratagene, La Jolla, CA). After induction of expression with isopropyl 1-thio-β-d-galactopyranoside, the bacteria were opened by sonification, and the Padron0.9 proteins were purified by Ni-NTA (Ni2+-nitrilotriacetate) affinity chromatography and subsequent size-exclusion chromatography according to standard procedures. During the whole purification procedure, the proteins were kept at 4 °C in buffered conditions (pH 7.5). The purified proteins were concentrated to ∼26 mg/ml by ultrafiltration and taken up in 20 mm Tris/HCl and 120 mm NaCl (pH 7.5) for crystallization. Padron0.9 was crystallized without removal of the N-terminal His6 tag by sitting drop vapor diffusion at room temperature (20 °C), by employing a reservoir of 35% (w/v) polyethylene glycol 400, 5% (w/v) polyethylene glycol 3000, 0.1 m Hepes (pH 7.5), 10% (w/v) glycerol, and 0.1 m spermidine. The measured pH of the crystallization solution was 6.6. Crystals appeared within 1 day and continued to grow for 2 weeks.
To switch whole Padron0.9 protein crystals into the on state, crystals were irradiated in the crystallization solution with blue light (488 ± 5 nm, 2.4 W/cm2) until the fluorescence reached a maximum. For the off state structure, crystals were first fully switched on and then irradiated with UV light (405 ± 5 nm, 5.3 W/cm2) until the fluorescence decreased to ∼5% of the initial value. After switching, care was taken to handle the crystals under red light illumination. Approximately 20 s after switching, the crystals were flash-frozen in liquid nitrogen. Diffraction data were collected at the PXII beamline of the Swiss Light Source (Villigen, Switzerland) at 100 K, using a MarResearch (Hamburg, Germany) CCD detector and processed with the HKL package (23). Both structures were solved essentially as described for Dronpa (11). For details, see supplemental Table 2.
Optical Switching
Photoswitching experiments were performed by using a custom-built, computer-controlled fluorescence microscope (Leica, Benzheim, Germany) equipped with a 40 × NA 0.6 air objective lens and two 100-W Hg lamps. For reversible switching, blue (488 ± 5 nm, 2.4 W/cm2) and UV (405 ± 5 nm, 5.3 W/cm2) light was used. Fluorescence intensities were recorded with a photomultiplier tube (HR9306-0; Hamamatsu, Hamamatsu, Japan) using a 500-nm long pass detection filter (HQ 500 LP; AHF Analysentechnik, Tübingen, Germany).
Protein Characterization
For the determination of the absorption, excitation, and fluorescence spectra, 2 μl of the protein solution was quantitatively transferred into the fluorescent or the nonfluorescent state by irradiation with blue light (488 ± 5 nm) or UV light (405 ± 5 nm), respectively, using a fluorescence microscope equipped with a 20 × air objective lens (N Plan 0.40 NA). The switching was monitored by measuring the fluorescence signal. After maximal switching, the proteins were diluted, and the absorption and emission spectra were immediately recorded with a Varian Cary 4000 UV/VIS spectrophotometer and a Varian Cary Eclipse fluorescence spectrophotometer (Varian, Palo Alto, CA), respectively. For pH titration experiments, the switched protein solution was diluted 50-fold into the appropriate buffer (pH 4–6, 0.1 m citrate buffer; pH 7 and 8, 0.1 m Tris-HCl; and pH 9 and 10, 0.1 m glycine buffer). To determine the fluorescence excitation spectra, the fluorescence was recorded at 525 nm. For the emission spectra, Padron0.9 was irradiated at 503 nm.
Free Energy Calculations
The difference in free energy for adding a proton to the anionic chromophore between the cis and trans configurations was determined by classical thermodynamic integration, combined with hybrid quantum/classical free energy perturbation. The x-ray structures of PadronOn and PadronOff were used as the starting coordinates. For details see supplemental “Materials and Methods.”
RESULTS
Spectral Properties of Padron0.9
Padron0.9 is an RSFP with positive switching characteristics (Fig. 1A and supplemental Table 1). It is a variant of Padron (12) differing at two amino acid positions (Y116C and K198I, all numbering according to the Dronpa sequence) on its surface (supplemental Fig. 1), slightly increasing its tendency for dimerization (supplemental Fig. 2), which facilitated crystallization. It differs from Dronpa by 10 amino acid residues, of which only two (V157G and M159Y) were required to reverse the switching characteristics.
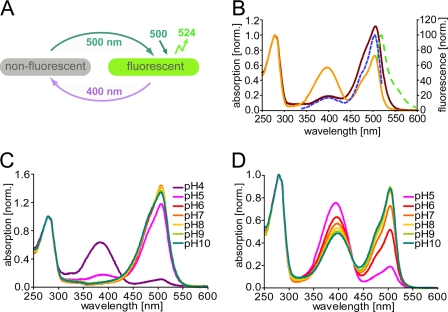
FIGURE 1.
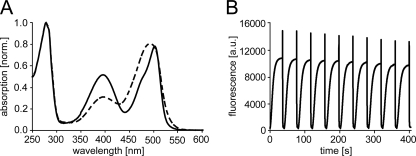
Spectroscopic properties of Padron0.9. A, schematic of the switching cycle. B, properties of purified protein at pH 7.0. Absorption spectrum of the nonfluorescent equilibrium state protein (solid dark red line). Absorption (solid orange line), fluorescence excitation (dashed blue line) and fluorescence emission (dashed green line) spectra of the protein in the fluorescent state. C, pH dependence of the absorption spectra in the nonfluorescent state. D, pH dependence of the absorption spectra in the fluorescent state.
At equilibrium, Padron0.9 adopts almost exclusively the nonfluorescent state, with a main absorption band at 504 nm (Fig. 1B and supplemental Table 1). Upon irradiation at this band, the nonfluorescent state is transferred into the fluorescent state, resulting in the emergence of an absorbance band at 395 nm and a decrease in the absorbance at 504 nm. Fluorescence emission, peaking at 524 nm, appears upon excitation of the 504 nm band of the fluorescent state. An excitation of the 395 nm band results only in weak fluorescence (Fig. 1B) and concomitantly transfers the protein to the nonfluorescent state.
The pH dependence of the absorption spectra reveals that the 504 nm band in the nonfluorescent and fluorescent states represents the deprotonated form of the chromophore (Fig. 1, C and D). The 395 nm band of the fluorescent state, which corresponds to the protonated form, is also observable upon titration of the nonfluorescent state to pH values <5.
In the nonfluorescent state, all reported Dronpa variants with negative switching characteristics have a largely protonated chromophore (10, 12). In contrast, in the nonfluorescent Padron0.9, the chromophore is almost exclusively in the deprotonated form, which is also the protonation state of the fluorescent chromophore. Hence, we conclude that protonation alone is insufficient to explain the absence of fluorescence.
Chromophore Structures of Padron0.9On and Padron0.9Off
To further investigate the inverted switching mechanism of Padron0.9, we crystallized the protein at a pH of 6.6 in the dark. Padron0.9 crystallized into octahedrally shaped crystals that could be reversibly switched at room temperature between a fluorescent form and a nonfluorescent form by alternating irradiation with blue (488 ± 5 nm) and blue and UV (405 ± 5 nm) light, the same wavelengths used to switch the protein in solution. Cycling of light-driven switching of the fluorescence of the crystal could be repeated with minimal loss in the maximum fluorescence signal (Fig. 2, A and B).
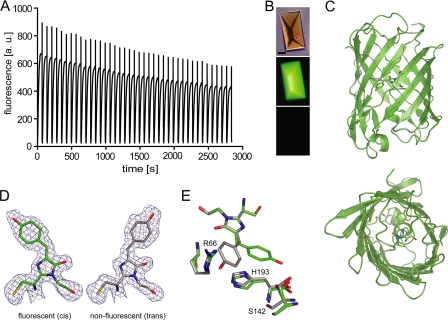
FIGURE 2.
Reversible photoswitching of Padron0.9. A, fluorescence signal of 40 switching cycles recorded on a single protein crystal in crystallization buffer (pH 6.6) at room temperature. Switching was performed by irradiation with blue light (488 ± 5 nm, 2.4 W/cm2, 66 s) alternating with UV light (405 ± 5 nm, 5.3 W/cm2, 4.5 s) together with blue light. B, images of the protein crystal. Top, bright field image. Middle and bottom, crystal in the fluorescent and nonfluorescent state, respectively. Scale bar, 20 μm. C, overlay of the on and the off structures displayed in two orthogonal views. D, chromophore conformations in the fluorescent (cis) and the nonfluorescent (trans) states (green, carbon fluorescent state; gray, carbon nonfluorescent state; red, oxygen; blue, nitrogen). Final 2Fo − Fc electron densities around the chromophores are contoured at the 1σ level. E, no pronounced amino acid rearrangements are observed in the x-ray structures upon photochromic switching. Note that Ser142 of Padron0.9On adopts two alternative conformations. a.u., arbitrary units.
To obtain the structure of Padron0.9 in the fluorescent state, crystals in solution were irradiated at room temperature with blue light (2.4 W/cm2) for 1–2 min until the fluorescence reached its maximum. For the nonfluorescent state structure, the crystals were first fully transferred into the fluorescent state and then irradiated with UV light (5.3 W/cm2) for ∼10 s until the crystals were almost nonfluorescent (<5% of the maximum fluorescence). For both structures, ∼20 s after irradiation, the crystals were flash-frozen in liquid nitrogen. This delay is three orders of magnitude shorter than the ensemble thermal relaxation half-time from the fluorescent into the nonfluorescent state (∼4 h, supplemental Table 1). Hence, we captured the proteins in the respective switched ground states. The x-ray structures of Padron0.9On (Protein Data Bank code 3LS3) and Padron0.9Off (Protein Data Bank code 3LSA) were refined to resolutions of 1.65 and 1.80 Å, respectively. The data collection and refinement statistics are compiled in supplemental Table 2.
The x-ray structures reveal that Padron0.9 adopts a classical β-can fold (Fig. 2C). Analogous to Dronpa in the fluorescent state, the Padron0.9 fluorescent (on) state chromophore adopts a cis conformation, whereas in the nonfluorescent (off) state, it is in a trans conformation. The well resolved electron densities for the hydroxyphenyl groups exclude the possibility of coexisting multiple chromophore conformations or a substantially disordered nonfluorescent ground state in the crystals at 100 K (Fig. 2D).
Similar to the situation in Dronpa, the imidazolinone ring of the Padron0.9 chromophore almost stays in place during the cis-trans isomerization, rotating by only ∼1.5 degrees (Fig. 2E). Therefore, the cis-trans isomerization at the methine bridge connecting the imidazolinone with the p-hydroxyphenyl ring results in an ∼7 Å movement of the apical hydroxyl group of the p-hydroxyphenyl ring. In Dronpa, this large movement is accommodated in the surrounding protein matrix by the movement of a number of nearby moving residues (17, 21). As described above, in Padron0.9, no comparable rearrangement of the surrounding protein matrix is seen in the x-ray structures (Fig. 2E).
Gly157, Tyr159, and Leu141 Are Crucial for the Switching Characteristics of Padron0.9
The amino acid residue exchanges V157G and M159Y are essential to convert the negative switching Dronpa into a positive switching variant (12). A comparison of the x-ray structures of Padron0.9Off and DronpaOff reveals the immediate implication of these mutations (Fig. 3A). In Padron0.9Off, Tyr159 stabilizes the chromophore by forming a hydrogen bond to the hydroxyl group of the p-hydroxyphenyl ring. This interaction, which is absent in Dronpa, is essential for the deprotonation of the Padron0.9off trans chromophore and for shifting the thermal equilibrium to the trans conformation. A glycine residue is required at position 157, because the larger valine at this position would not allow Tyr159 to occupy its position at the trans site of the chromophore.
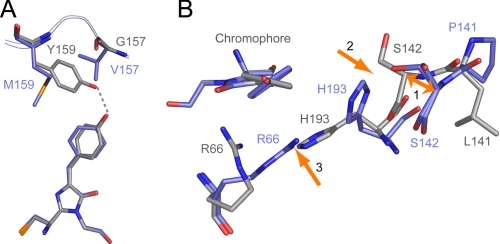
FIGURE 3.
Differences between Padron0.9Off and DronpaOff. Carbon atoms of Padron0.9Off are depicted in gray, those of DronpaOff in blue. A, V157G and M159Y are the key amino acid changes that convert the negative switching Dronpa into a positive switching variant. In Padron0.9, Tyr159 stabilizes the chromophore in the trans conformation. B, P141L rearranges the backbone of β-strand 7 (arrow 1), which impedes major structural rearrangements of the residues close to the Padron0.9 chromophore upon switching, in contrast to the situation in Dronpa (arrows 2 and 3). The lack of residue rearrangements in Padron0.9Off ultimately results in a more pronounced twisting of the p-hydroxyphenyl ring of the trans chromophore as compared with DronpaOff. For details on the arrows, see main text.
Furthermore, in Padron0.9Off, Leu141 (instead of the Pro141 in Dronpa) results in a shift of the relative position of Ser142 (the Cα atom of Ser142 is shifted by ∼2 Å) into the direction of the cis chromophore pocket, as compared with DronpaOff (Fig. 3B, arrow 1). As a result, in Padron0.9, His193Off apparently cannot move into a cavity which is available in DronpaOff (Fig. 3B, arrow 2), thereby forcing Arg66Off to stay at its place (Fig. 3B, arrow 3). The static Arg66 forces the p-hydroxyphenyl ring of Padron0.9Off to rotate, resulting in an off state chromophore with a marked torsion. As a consequence, no amino acid rearrangements are required to accommodate the cis-trans isomerization in Padron0.9. Hence the lack of residue rearrangement can be traced to the P141L exchange.
Properties of the Fluorescent and the Nonfluorescent State
We found that off state Padron0.9 cooled to ∼170 K does not exhibit a significant increase in fluorescence, indicating that chromophore flexibility is not a major channel for radiation-less decay of the excited Padron0.9 trans chromophore. This is also supported by the observation that in the protein crystal, the nonfluorescent Padron0.9 trans chromophore is well stabilized, participating in altogether 8–9 hydrogen bonds to nearby residues, three water-mediated hydrogen bonds, and numerous van der Waals interactions. A comparable number of interactions are observed for the fluorescent cis chromophore (supplemental Fig. 3). In fact, in Padron0.9, the fluorescent and the nonfluorescent states exhibit average root mean square deviations of 0.45 Å for the atoms of the immediate chromophore surroundings. The respective values for the chromophore environments of the four crystallographically independent molecules within the same crystal and state are very similar (on state, 0.50 Å; off state, 0.32 Å). Thus, the cis and the trans chromophores are attached to the surrounding protein matrix to a comparable degree (supplemental Fig. 3). Hence, in Padron0.9, the major structural differences between the fluorescent and the nonfluorescent states are the conformations and torsions of the chromophore.
Protonation of the Chromophore and Its Environment
The cis and the trans chromophore are differently protonated (Fig. 1). In fluorescent proteins, it is very challenging to determine the protonation state of the amino acid residues neighboring the chromophore experimentally. To address this issue and to clarify how the protein environment in Padron0.9 controls the protonation state of the chromophore in the cis and the trans configuration, we performed molecular dynamics-based free energy calculations on the Padron0.9On and Padron0.9Off structures. Details of the simulations are provided in the supplemental “Materials and Methods.” We found a difference in protonation-free energy of 11.4 ± 3 kJ/mol between the trans and cis chromophore. Thus, the free energy calculations predict that isomerization from cis to trans lowers the pKa of the chromophore by 2.0 ± 0.5 pKa. This trend is in good agreement with the experimentally determined pKa shift of ∼ 1.5 (pKaOff ∼ 4.5; pKaOn ∼ 6.0) (Fig. 1, C and D).
In addition, our calculations provide information about the protonation states of titratable residues neighboring the chromophore, namely Glu144, His193, and Glu211. We performed the free energy computations with four possible different protonation states of these residues (Fig. 4 and supplemental Table 4). We find that only if the imidazolinone ring of His193 is protonated at the Nδ position and if the side chain of His193 donates a hydrogen bond to the Glu211 side chain while accepting a hydrogen bond from the protonated Glu144 side chain (Fig. 4A), the pKa shift is in agreement with the experimentally determined absorption data. All other analyzed possibilities (Fig. 4, B–D) result in pKa shifts in the opposite direction (supplemental Table 4) and are thus not consistent with the experimental data.
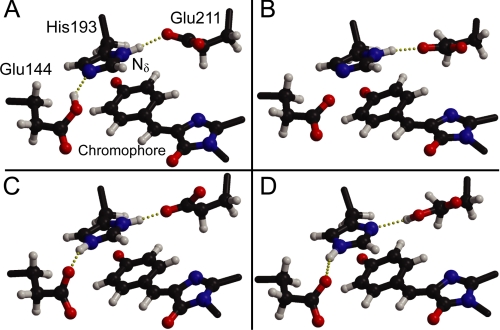
FIGURE 4.
Alternative protonation states in the chromophore pocket of Padron0.9. Depicted are snapshots of molecular dynamics simulations. A, singly protonated His193, donating a hydrogen bond to Glu211 and accepting a hydrogen bond from Glu144. B, singly protonated His193, and both glutamates deprotonated. C, doubly protonated His193, donating hydrogen bonds to Glu144 and Glu211. D, singly protonated His193, accepting a hydrogen bond from Glu211 and donating a hydrogen bond to Glu144. The situation without a shared proton between His193 and Glu211 was ruled out on the basis of the x-ray structure and not considered further. Free energy computations reveal that alternative (A) represents the actual situation in the protein.
Distinct Absorption Cross-sections in the Fluorescent and the Nonfluorescent State Are Not Essential for Efficient Switching
In Padron0.9, as in all other published RSFPs so far (12, 13, 24), the cis and the trans chromophores have clearly different absorption spectra due to different protonation equilibria in the two states. This observation led to the suggestion that different absorption cross-sections for the switching wavelengths in the fluorescent and the nonfluorescent states are essential for effective switching in RSFPs. In contrast, upon analysis of numerous Padron0.9 variants, we identified a variant demonstrating that effective switching is determined by the probabilities of isomerization rather than by different cross-sections for the switching wavelengths. We found that reverting Leu141 to a proline residue results in the emergence of a strong absorbance band at 395 nm, corresponding to a protonated chromophore in the nonfluorescent state (Fig. 5A). As a result, the absorption spectra of the fluorescent and the nonfluorescent states of Padron0.9-L141P are largely similar. Here, the ratio between the absorption cross-sections for 405 nm and 488 nm in the fluorescent and nonfluorescent states are 1:1.2 and 1:2.7, respectively (Fig. 5A). In comparison, in Dronpa, these ratios are 1:18.1 and 17.2:1, respectively (10). Nonetheless, Padron0.9-L141P is readily and reversibly switchable and can be switched off to 2.9% of the maximum fluorescence (Fig. 5B and supplemental Table 1). Thus, the ensemble switching efficiency does not depend on the absorption cross-sections per se, which are predominantly determined by the protonation state of the chromophore. Rather, the tendency to isomerize depends on the underlying potential energy surfaces in the excited state, which are distinct in the cis and the trans chromophores.
FIGURE 5.
Properties of Padron0.9-L141P. A, absorption spectra in the fluorescent (solid) and the nonfluorescent (dashed) states, recorded on purified proteins at pH 7.5. In contrast to Padron0.9, the nonfluorescent state of Padron0.9-L141P exhibits a strong absorption band at 395 nm, corresponding to the protonated chromophore. B, fluorescence signal recorded upon reversible switching of Padron0.9-L141P in solution by alternating irradiation with blue light (488 ± 5 nm, 2.4 W/cm2, 36 s) or UV light (405 ± 5 nm, 5.3 W/cm2, 4.5 s) together with blue light. a.u., arbitrary units; norm., normalized.
DISCUSSION
Although in most fluorescent proteins the chromophore adopts a cis conformation, there are exceptions exhibiting a trans chromophore (25). Thus, the conformation of the chromophore per se is insufficient to explain the difference in fluorescence between the on and the off states. This raises the question why the cis chromophore in Padron0.9 is fluorescent, as in all other currently described RSFPs, whereas the trans chromophore is not.
Crystallographic studies capture the chromophore in the respective ground states and are not meant to investigate the short-lived excited states that might be partially disordered. Presumably and not unexpectedly, the photochromic proteins may exhibit flexibility in the excited off state (22). It may be noted, however, that the cooling off state Padron0.9 to ∼170 K does not increase the fluorescence noticeably. Therefore, it appears unlikely that the lack of fluorescence can solely be attributed to chromophore flexibility, but rather that the torsion of the trans state chromophore is a major factor determining the absence of fluorescence.
From detailed studies on organic stilbenes, it is known that planar chromophoric systems are more fluorescent than those with a strong torsion (26). Strikingly, in Padron0.9, the angle θ spanned by the planes of the chromophoric five- and six-membered rings is almost identical in both the fluorescent and nonfluorescent ground states (supplemental Fig. 4). Consequently, co-planarity per se appears not to be an appropriate measure to distinguish fluorescent from nonfluorescent chromophores.
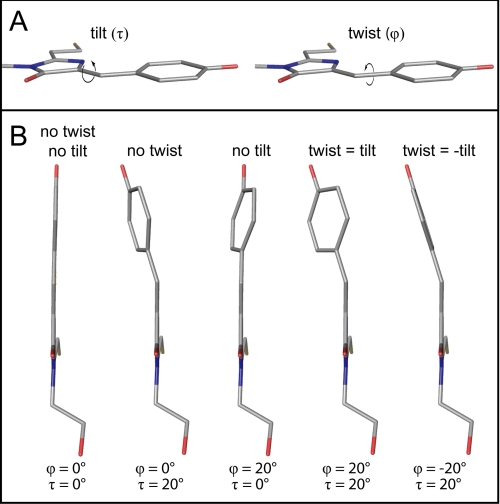
However, the position of the two chromophoric rings relative to each other is not unambiguously described by the angle θ. Rather, the “tilt” (τ) and “twist” (ϕ) angles (Fig. 6A) fully describe the torsion of the chromophoric system. If τ and ϕ have the same direction, the torsion (“propeller twist”) within the chromophore is increased, whereas if τ and ϕ have opposite directions, the torsion is reduced, and the chromophoric system is more smoothly bent (Fig. 6B). A chromophore with a smooth bend, as opposed to a pronounced propeller twist, may be achieved even in a chromophore with large τ and ϕ angles. Therefore, the modulus of the sum of τ and ϕ, rather than the sum of the modulus of τ and the modulus of ϕ, is an appropriate measure for the bending of the chromophore.
FIGURE 6.
The modulus of the sum of the dihedral angles τ and ϕ is a measure for the torsion between the two chromophoric rings. A, definition of the tilt (τ) and the twist (ϕ) angle in the Padron0.9 chromophore. B, hypothetical Padron0.9 chromophores with different values for τ and ϕ. A fully planar chromophore is only achieved when both τ and ϕ are 0° (far left chromophore). However, the chromophore adopts a smooth bend without torsion when τ and ϕ have the same values but opposing directions (far right chromophore).
Indeed, by analyzing all available x-ray structures of photochromic FPs, we find that the modulus of the sum of τ and ϕ is always smaller in the fluorescent state than in the nonfluorescent state (supplemental Table 5). This rule holds true even for chromophore structures of Dronpa and one of its variants, which were predicted by molecular dynamics simulations (21). Hence, the modulus of the sum of τ and ϕ appears to be a robust measure to predict the tendency of a green fluorescent protein-like chromophore to fluoresce.
The importance of this factor may vary between different fluorescent proteins. In Padron0.9, the modulus of the sum of τ and ϕ is 11.6 ± 1.1 and 22.3 ± 2.1 for the fluorescent and the nonfluorescent states, respectively (supplemental Table 5). The same tendency is observed in the RSFP asFP595-A143S, although the sums of τ and ϕ in both states of the chromophore are only slightly different (10.3 ± 1.5 and 14.7 ± 2.3, respectively). Intriguingly, for this protein, it has been shown that the cis-oriented chromophore is much better stabilized by the surrounding protein matrix than the trans-oriented chromophore (15), further supporting the view that the rigidity of the chromophoric system is another key factor determining its ability to fluoresce.
Comparatively little is known about the proton dynamics within the β-barrel and the protonation states of titrable amino acids residues close to the chromophore. Our molecular dynamics simulations of Padron0.9 indicate that no changes in the overall protonation pattern were required to obtain the pKa shift of two units. We therefore suggest that the proton for neutralizing the chromophore is taken up from the bulk solution, rather than from somewhere in the protein. In this case, an equilibrium between the neutral and anionic forms of the chromophore would be established after the photoisomerization.
We found that the titration behavior of on state Padron0.9 is highly irregular and exhibits a complicated pH dependence (Fig. 1D). Increasing the pH above 6.0, the estimated pKa of the cis chromophore does not result in a complete disappearance of the 395 nm absorbance band associated with the protonated species. An irregular titration behavior suggests that there are strong interactions between groups with similar intrinsic pKa values (27).
Thus, we assume a coupling between the Padron0.9 cis chromophore (on state) with an other residue with a similar pKa. Potential interaction partners of the cis chromophore are the ionizable groups of cysteines (pKa ∼ 8.55) or histidines (pKa ∼ 6.54) (28). To identify a potential coupling amino acid residue, we first mutated (in computer simulations as well as experimentally) Cys62 and Cys171 individually against serines, whose hydroxyl groups are normally unreactive and would not get deprotonated. Both free energy calculations (supplemental Table 6) as well as pH titration experiments (supplemental Fig. 5) did not modify the titration behavior of the on state. Therefore, these data exclude Cys62 or Cys171 as the primary cause of the peculiar pH titration behavior of on state Padron0.9.
The other residue that might induce the irregular titration behavior of the chromophore is His193, which forms a π-stack with the hydroxyphenyl ring of the cis chromophore. Indeed, our molecular dynamics simulations showed that deprotonating His193 (together with Glu144) results in an upward shift of the pKa by ∼3 units (i.e. the protonation free energy decreases from 1105.8 to 1122.4 kJmol−1; supplemental Table 4), indicating a strong coupling between His193 and the chromophore. We therefore suggest that the irregular titration behavior of the cis chromophore of Padron0.9 may be mediated by a coupling of His193 with the chromophore, stabilizing the protonated chromophore at high pH values.
In summary, we demonstrated in this study by analysis of Padron0.9-L141P that distinct absorption cross-sections for switching in the on and the off states are not necessary for efficient light-driven reversible switching. We analyzed the molecular basis for the antipodal switching characteristics of Padron0.9 and Dronpa. Although in Padron0.9, in contrast to Dronpa, no further rearrangements of the protein matrix occur due to the single mutation P141L, the essential element of switching is a light-induced cis-trans isomerization of the chromophore.
Supplementary Material
Acknowledgments
We thank the staff of beamline PXII of the Swiss Light Source (Villigen, Switzerland) for support during diffraction data collection. We also thank S. Löbermann for excellent technical assistance, D. Schwarzer for insightful discussions, and J. Jethwa for carefully reading the manuscript.
This work was supported by the Gottfried Wilhelm Leibniz Program of the Deutsche Forschungsgemeinschaft (to S. W. H.).
The atomic coordinates and structure factors (codes 3LSA and 3LS3) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” Tables 1–6, Figs. 1–6, and additional references.
- FP
- fluorescent protein
- RSFP
- reversibly switchable FP
- W
- watt(s).
REFERENCES
- 1.Tsien R. Y. (1998) Annu. Rev. Biochem. 67, 509–544 [DOI] [PubMed] [Google Scholar]
- 2.Chudakov D. M., Lukyanov S., Lukyanov K. A. (2005) Trends Biotechnol. 23, 605–613 [DOI] [PubMed] [Google Scholar]
- 3.Fernández-Suárez M., Ting A. Y. (2008) Nat. Rev. Mol. Cell Biol. 9, 929–943 [DOI] [PubMed] [Google Scholar]
- 4.Day R. N., Davidson M. W. (2009) Chem. Soc. Rev. 38, 2887–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nienhaus G. U., Wiedenmann J. (2009) Chemphyschem 10, 1369–1379 [DOI] [PubMed] [Google Scholar]
- 6.Sample V., Newman R. H., Zhang J. (2009) Chem. Soc. Rev. 38, 2852–2864 [DOI] [PubMed] [Google Scholar]
- 7.Lukyanov K. A., Chudakov D. M., Lukyanov S., Verkhusha V. V. (2005) Nat. Rev. Mol. Cell Biol. 6, 885–891 [DOI] [PubMed] [Google Scholar]
- 8.Lippincott-Schwartz J., Patterson G. H. (2009) Trends Cell Biol. 19, 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukyanov K. A., Fradkov A. F., Gurskaya N. G., Matz M. V., Labas Y. A., Savitsky A. P., Markelov M. L., Zaraisky A. G., Zhao X., Fang Y., Tan W., Lukyanov S. A. (2000) J. Biol. Chem. 275, 25879–25882 [DOI] [PubMed] [Google Scholar]
- 10.Ando R., Mizuno H., Miyawaki A. (2004) Science 306, 1370–1373 [DOI] [PubMed] [Google Scholar]
- 11.Stiel A. C., Trowitzsch S., Weber G., Andresen M., Eggeling C., Hell S. W., Jakobs S., Wahl M. C. (2007) Biochem. J. 402, 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andresen M., Stiel A. C., Fölling J., Wenzel D., Schönle A., Egner A., Eggeling C., Hell S. W., Jakobs S. (2008) Nat. Biotechnol. 26, 1035–1040 [DOI] [PubMed] [Google Scholar]
- 13.Henderson J. N., Ai H. W., Campbell R. E., Remington S. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6672–6677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stiel A. C., Andresen M., Bock H., Hilbert M., Schilde J., Schönle A., Eggeling C., Egner A., Hell S. W., Jakobs S. (2008) Biophys. J. 95, 2989–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andresen M., Wahl M. C., Stiel A. C., Gräter F., Schäfer L. V., Trowitzsch S., Weber G., Eggeling C., Grubmüller H., Hell S. W., Jakobs S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13070–13074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schüttrigkeit T. A., von Feilitzsch T., Kompa C. K., Lukyanov K. A., Savitsky A. P., Voityuk A. A., Michel-Beyerle M. E. (2006) Chem. Phys. 323, 149–160 [Google Scholar]
- 17.Andresen M., Stiel A. C., Trowitzsch S., Weber G., Eggeling C., Wahl M. C., Hell S. W., Jakobs S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13005–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fron E., Flors C., Schweitzer G., Habuchi S., Mizuno H., Ando R., Schryver F. C., Miyawaki A., Hofkens J. (2007) J. Am. Chem. Soc. 129, 4870–4871 [DOI] [PubMed] [Google Scholar]
- 19.Schäfer L. V., Groenhof G., Klingen A. R., Ullmann G. M., Boggio-Pasqua M., Robb M. A., Grubmüller H. (2007) Angew. Chem. Int. Ed. 46, 530–536 [DOI] [PubMed] [Google Scholar]
- 20.Adam V., Lelimousin M., Boehme S., Desfonds G., Nienhaus K., Field M. J., Wiedenmann J., McSweeney S., Nienhaus G. U., Bourgeois D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18343–18348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moors S. L. C., Michielssens S., Flors C., Dedecker P., Hofkens J., Ceulemans A. (2008) J. Chem. Theory Comput. 4, 1012–1020 [DOI] [PubMed] [Google Scholar]
- 22.Mizuno H., Mal T. K., Wälchli M., Kikuchi A., Fukano T., Ando R., Jeyakanthan J., Taka J., Shiro Y., Ikura M., Miyawaki A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9227–9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24.Ando R., Hama H., Yamamoto-Hino M., Mizuno H., Miyawaki A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12651–12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiedenmann J., Schenk A., Röcker C., Girod A., Spindler K. D., Nienhaus G. U. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11646–11651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oelgemöller M., Brem B., Frank R., Schneider S., Lenoir D., Hertkorn N., Origane Y., Lemmen P., Lexe J., Inoue Y. (2002) J. Chem. Soc. Perkin Trans. 2, 1760–1771 [Google Scholar]
- 27.Klingen A. R., Bombarda E., Ullmann G. M. (2006) Photochem. Photobiol. Sci. 5, 588–596 [DOI] [PubMed] [Google Scholar]
- 28.Thurlkill R. L., Grimsley G. R., Scholtz J. M., Pace C. N. (2006) Protein Sci. 15, 1214–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.