Fig. 3.
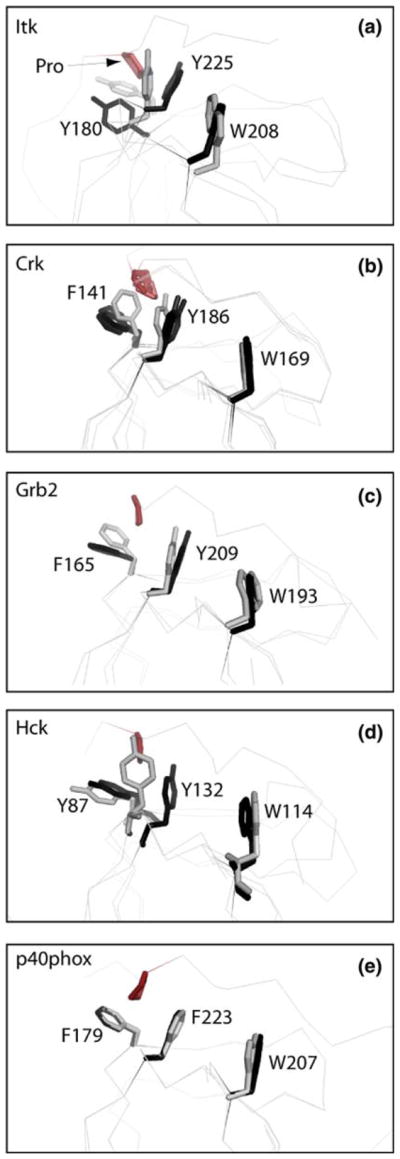
Superpositions of SH3 domain structures either bound to a proline-rich peptide ligand (colored black) or unbound (colored gray). The bound and unbound structures for each SH3 domain were superimposed using all SH3 domain atoms. Three aromatic residues within the SH3 binding pocket are depicted (Y180, Y225 and W208 for Itk) and labeled for each structure. Residue labels correspond to the full length numbering for each protein. For each bound structure the proline from the ligand that contacts Y180 and Y225 (Itk numbering) is indicated in red. The pdb code for each free and bound structure is: (a) Itk: this study (free), 1awj (bound); (b) c-Crk: 1m30 (free), 1bo7 (bound), 1cka (bound), 1ckb (bound); (c) Grb2: 1gfc (free), 1io6 (bound); (d) Hck: 4hck (free), 2oj2 (bound); (e) p40phox: 1w6x (free), 1w70 (bound)
