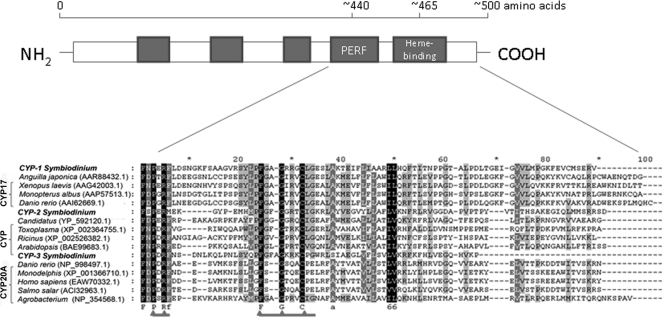
FIG. 2.
Diagram of the cytochrome P450 primary structure (48), showing the distribution of the major conserved domains (filled boxes), and multiple-sequence alignment of the Symbiodinium CYP C-terminal regions, including the conserved PERF domain (PXRF) and heme-binding region (FXXGXRXCXG), constructed with Clustal W. Alignment was performed using the following pairwise alignment options: Blossum30 protein weight matrix, a gap-opening penalty of 10, and a gap extension penalty of 0.1. Predicted amino acid sequences encoded by the Symbiodinium ESTs designated CYP-1, CYP-2, and CYP-3 were aligned with the selected sequences producing the best hits according to a BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The identical residues in all sequences are indicated; amino acids conserved in 100% of the sequences are indicated by white letters with a black background, amino acids conserved in >80% of the sequences are indicated by white letters with a gray background, and amino acids conserved in >60% of the sequences are indicated by black letters with a gray background.