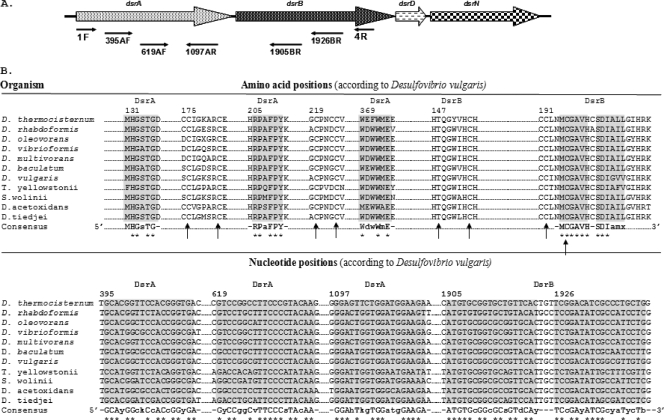
FIG. 1.
(A) dsrABDN operon of Desulfovibrio vulgaris. The locations of the primers designed in this study and that of the known primers DSR1F and DSR4R (54) are indicated. (B) Alignment fragments for the priming sites on DsrAB (∼750 amino acids) and the dsrA and dsrB genes (∼1,900 nucleotides). Uppercase letters in the consensus sequence indicate the unit fully conserved at the position among the SRP analyzed. Lowercase letters in the consensus correspond to designated ambiguities. The consensus sequence for the reverse primer was processed for proper orientation. The complete [Fe4S4]-seroheme binding site motif (Cys/Thr-X5-Cys)-Xn-(Cys-X3-Cys) is indicated by arrows. The dsrA and dsrB genes shown in the alignments are from Desulfotomaculum thermocisternum DSM10259T (AF074396), Desulfobulbus rhabdoformis DSM8777T (AJ250473), Desulfobacterium oleovorans (AF482464), Desulfobacter vibrioformis DSM8776T (AJ250472), Desulfococcus multivorans DSM2059T (U58126), Desulfomicrobium baculatum (AF4282460), Desulfovibrio vulgaris DSM644T (U16723), Thermodesulfovibrio yellowstonii DSM 11347, Syntrophobacter wolinii DSM 2805 (AF418192), Desulfobacca acetoxidans (AF482453), and Desulfomonile tiedjei DSM 6799 (AF334595).