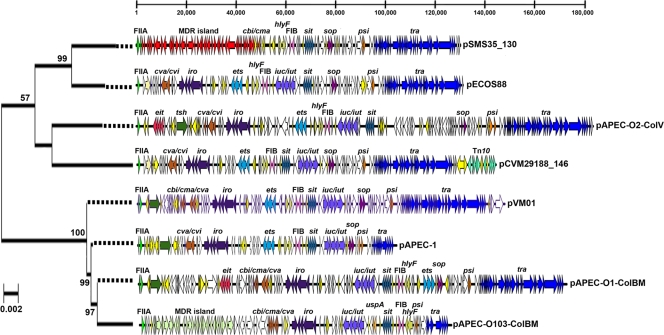
FIG. 2.
Linear maps of sequenced ColV and ColBM plasmids. The bar at the top indicates positions (in base pairs). The maps begin with RepFIIA and are drawn to scale. Virulence genes of interest are identified and indicated by different colors. The plasmids are ordered according to evolutionary relationships of concatenated gene sequences (hlyF, traX, finO, repA, repA1). The evolutionary history was inferred using the neighbor-joining method (54). Bootstrap consensus trees were inferred using 500 replicates. Bootstrap confidence values greater than 50% are indicated at the nodes. The tree is drawn to scale based on evolutionary distances, and dotted lines extend the branches to the plasmid maps. Distances were computed using the maximum composite likelihood method (scale bar = 0.02 base substitution per site). Phylogenetic analyses were conducted with MEGA4 (55).