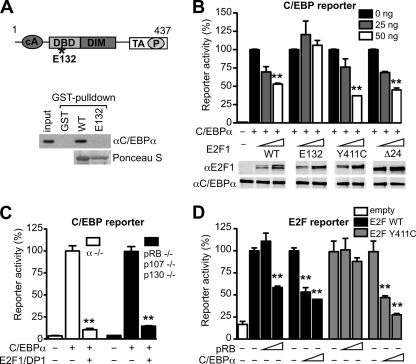
FIG. 3.
Cross-repression of C/EBPα and E2F1 is independent of pocket proteins. (A) At the top is a schematic representation of E2F1. cA, cyclin A-binding domain; DBD, DNA-binding domain; DIM, dimerization domain; TA, transactivation domain; P, pocket protein interaction domain; asterisk, approximate location of the E132 mutations. Indicated below are the results of GST-pulldown assays performed to assess the binding of C/EBPα (expressed in 293T cells) to either E2F1 WT or E2F1 E132 (GST fusion proteins). Bound proteins were detected by immunoblotting, and pulldown of GST proteins was controlled by Ponceau S staining. (B) An intact DNA-binding domain of E2F1 is required to repress the transcriptional activity of C/EBPα. Different E2F1 mutants were tested for their ability to inhibit C/EBPα-mediated transcription: E132 (DNA binding deficient), Y411C (pRB binding deficient), and Δ24 (cyclin A binding deficient). 293T cells were transfected with a C/EBP responsive gene reporter, C/EBPα (100 ng), and with E2F1 mutant expression constructs, as indicated (E2F1 is always in combination with 50 ng of HA-DP1). The dose-dependent effects of E2F1 mutants were tested. Protein expression was controlled by immunoblotting, and the lanes were run on the same gel, but the image was divided into different panels for the shake of simplicity (lower panels). **, P < 0.01 versus C/EBPα alone, which was set to 100. (C) E2F1-mediated repression of C/EBPα is independent of pocket proteins. C/EBPα−/− MEFs or pRB−/− p107−/− p130−/− MEFs were transfected with C/EBP-responsive gene reporter, C/EBPα, HA-DP1, and HA-E2F1 (all at 100 ng). **, P < 0.01 versus C/EBPα alone, which was set to 100. (D) C/EBPα-mediated repression of E2F is independent of pocket proteins. pRB−/− p107−/− p130−/− MEFs were transfected with E2F-responsive gene reporter (100 ng) and with HA-DP1 (50 ng), in combination with 50 ng of either E2F1 WT or E2F1 Y411C expression constructs. Increasing amounts of pRB or C/EBPα (triangle: 100 or 200 ng) were also transfected as indicated to assess their repression on E2F1. As a control, cells were transfected with empty vector. **, P < 0.01 versus E2F1-DP1 alone, which was set to 100. Reporter assays were performed in duplicate, plotted as the means ± the SEM. The data are representative of at least three independent experiments.