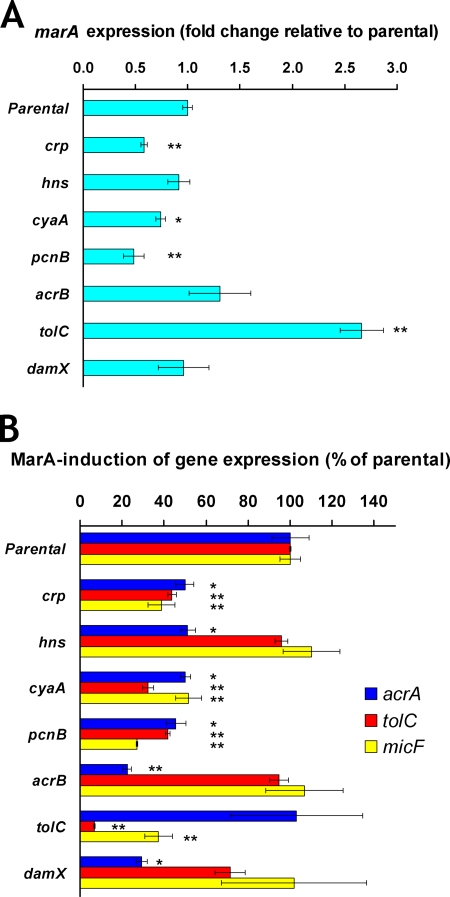
FIG. 2.
Gene expression in mutants with reduced MarA-mediated MDR to all antibiotics tested. The effect of each inactivated gene on the expression of marA, acrA, tolC, micF, and gapA was measured by RT-qPCR (see Materials and Methods). The results are presented as the average ± the standard error of the mean (n = 3 to 5). Statistically significant differences for a mutant compared to the level for the parental strain are shown as * (P < 0.05) or ** (P < 0.01). (A) Fold change in marA expression in different ΔmarR-derivative mutants relative to the level for the ΔmarR parental strain CR1000. Fold 1 means no change in marA expression in a mutant compared to the level for the parental strain. gapA was used as an endogenous reference gene, and its expression was not affected by marA (gapA expression was the same in the ΔmarR and ΔmarRA parental strains; data not shown). Except for pcnB, none of the inactivated genes tested significantly affected the expression of gapA. The pcnB mutant showed reductions in both gapA and marA levels. However, such reductions seem to be an effect specific for these genes and not a general effect produced by pcnB inactivation on all RNAs, since inactivation of pcnB in the ΔmarRA background produced the same reduction in gapA levels but did not decrease the levels of the other genes studied (acrA, tolC, and micF) (data not shown). (B) Effect of inactivation of each gene on MarA induction of acrA, tolC, and micF expression. The percentage of MarA induction of gene expression for each mutant was calculated by comparing the ΔmarR/ΔmarRA ratio (in terms of the number of tran-scripts per ng of RNA) observed for the mutant to that observed for the parental strain, as explained for the percentage of MarA-mediated MDR in Table 3, footnote a.