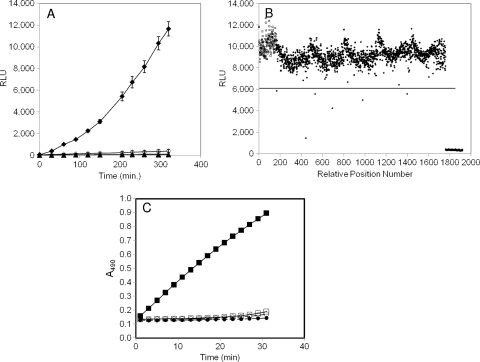
FIG. 1.
Characterization of bioluminescent and chromogenic reporter strains for identification of T3SS inhibitors. (A) Luminescence (in relative light units, RLU) from a chromosomal transcriptional fusion of exoT to the P. luminescens luxCDABE operon in wild-type (strain MDM852) or ΔpscC (strain MDM1355) P. aeruginosa PAO1 cells. Overnight cultures were diluted at time zero to A600 ∼ 0.025 and induced with 5 mM EGTA or left uninduced. RLU values were measured in 96-well opaque microplates throughout a 320-min time course. ⧫, MDM852 with EGTA; ⋄, MDM852 without EGTA; ▴, MDM1355 with EGTA; ▵, MDM1355 without EGTA. (B) Luminescence (RLU) from five 384-well microplates containing reporter strain MDM852 in a high-throughput screen for T3SS inhibitors. RLU values are shown at 300 min for 160 negative controls (□; fully induced by EGTA) in positions 1 to 160, for 160 positive controls (▴; no induction by EGTA) in positions 1,761 to 1,920, and for 1,600 samples (•) in positions 161 to 1,760. Six samples were designated hits because their RLU values displayed Z scores of >4 (i.e., >4 standard deviations below the average sample value, denoted as a horizontal line at 6,084 RLU). Compound 1 at position 443 was the most potent hit (Z score = 10) (Table 3). (C) Detection of ExoS′-βLA secretion from P. aeruginosa strains MDM973 (PAK) and MDM974 (PAK ΔpscC) carrying pUCP24GW-lacIQ-lacPO-exoS′-blaM, as measured by the hydrolysis of nitrocefin. A490 values are plotted versus time for MDM973 in the presence (▪) and absence (□) of 5 mM EGTA and for strain MDM974 in the presence (•) and absence (○) of 5 mM EGTA.