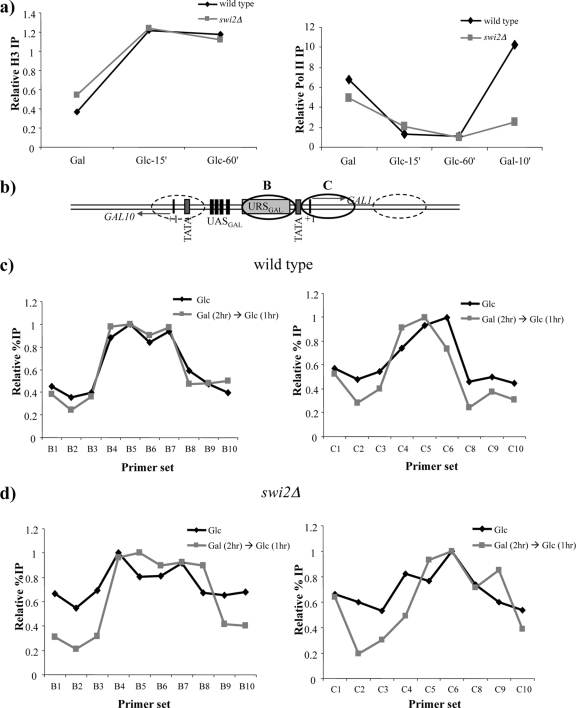
FIG. 5.
SWI/SNF promotes RNA polymerase II loading but does not generate alternate nucleosome positions at the GAL1 promoter. (a, left) Histone H3 ChIP of the wild-type and swi2Δ strains to measure nucleosome occupancy at the GAL1 promoter. The loss of SWI/SNF does not inhibit the kinetics of promoter nucleosome reloading during glucose repression. (Right) RNA polymerase II ChIP of the wild-type and swi2Δ strains showing that the faster recruitment during GAL1 reinduction is dependent on SWI/SNF. RNA polymerase II and H3 levels were tested at the GAL1 promoter and normalized to a telomere sequence (Chr VI, 70 bp from the right end). (b) Schematic representation of the GAL1-10 regulatory region. UASGAL marks the Gal4p binding sites. URSGAL is the binding site for the glucose-dependent repressor Mig1p. Ovals represent previously mapped nucleosome positions. Ovals shown as solid lines represent GAL1 promoter nucleosomes that are mapped in c and d. TATA represents the TBP binding sites, and +1 represents the transcription start sites. (c) Nucleosome-scanning ChIP with histone H3 antibody in the wild-type strain for promoter nucleosomes B (left) and C (right). Black lines represent cultures grown in glucose overnight, and gray lines represent short-term (1-h) glucose-repressed cultures following a brief GAL1 induction. On the x axes of graphs, B1 to B10 represent 10 primer pairs spanning positions −302 to +3 from the translation start site. C1 to C10 represent nine primer pairs spanning positions −148 to +160 from the translation start site. On the y axis, the relative percent IP of H3 normalized to a maximum value of 1 is plotted. (d) Same as panel c but with an swi2Δ strain.