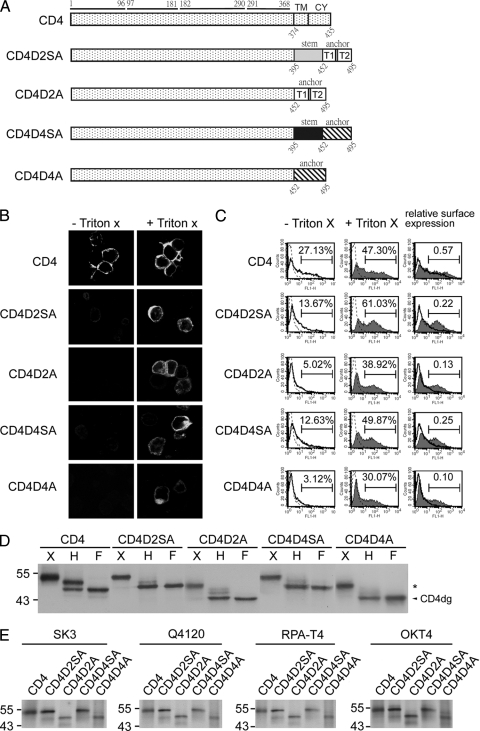
FIG. 1.
CD4 and chimeric CD4 constructs, surface expression, glycosylation pattern, and recognition by different MAbs. (A) Schematic diagram of the CD4 construct (pCB-CD4) and chimeric CD4 constructs containing the stem-TMD of DENV2 (CD4D2SA) or DENV4 (CD4D4SA) or TMD of DENV2 (CD4D2A) or DENV4 (CD4D4A). Open and gray bars indicate DENV2 sequences, and hatched and black bars indicate DENV4 sequences. The amino acid positions of the four domains of CD4 are shown above, and those of the stem, TMD, or CY domain are shown below. (B) 293T cells transfected with each of these constructs were subjected to indirect immunofluorescence assay using mouse anti-CD4 MAb RPA-T4 as described in Materials and Methods. (C) 293T cells transfected as described for panel A were permeabilized with or without Triton X-100 and subjected to flow cytometry analysis using MAb RPA-T4. Solid lines indicate MAb without Triton X-100, and solid lines with a shaded area indicate MAb with Triton X-100. Dotted lines indicate isotype controls, which overlap with those of mock-transfected cells (not shown). The relative surface expression of CD4 or chimeric CD4 was the ratio of the percentage of RPA-T4-positive cells in the absence of Triton X-100 to that in the presence of Triton X-100. (D) Lysates derived from 293T cells transfected as described for panel A were treated with endo H (H) or PNGase F (F) and subjected to Western blot analysis using RPA-T4. The arrowhead indicates deglycosylated CD4 (CD4dg), and asterisks indicate endo H-resistant bands. One representative experiment of three is shown. (E) 293T cells transfected as described for panel A were labeled with [35S]methionine at 20 h posttransfection and subjected to immunoprecipitation with anti-CD4 MAbs (RPA-T4, SK3, Q4120, and OKT4) and 12% PAGE as described in Materials and Methods. Molecular mass markers are shown in kDa.