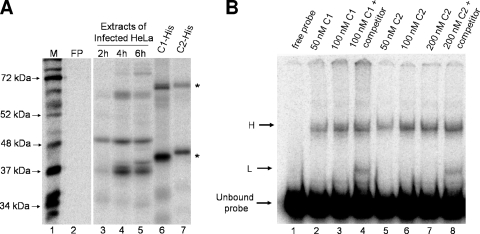
FIG. 4.
Binding of hnRNP C to the 5′ end of poliovirus negative-strand RNA under nondenaturing conditions. (A) Binding of recombinant hnRNP C to poliovirus negative-strand RNA. UV cross-linking assays were carried out using 50 μg of extracts generated from poliovirus-infected HeLa cells at 2 h (lane 3), 4 h (lane 4), and 6 h (lane 5) postinfection. Lanes 6 and 7 show UV cross-linked complexes with bacterially expressed His-tagged hnRNP C recombinant proteins containing a carboxyl-terminal hexahistidine tag. Lane 6, 0.5 μg hnRNP C1-His; lane 7, 0.5 μg of hnRNP C2-His. Image analysis was carried out using a phosphorimager. Lane M, marker proteins ([35S]methionine-labeled in vitro translation of poliovirus virion RNA). FP, free probe in the absence of any extract, as described in the legend to Fig. 2. (B) RNA mobility shift assays were carried out as described in Materials and Methods. Recombinant purified hnRNP C1 (lanes 2 to 4) or hnRNP C2 (lanes 5 to 8) was incubated with radiolabeled 5′(−) RNA probe. The competition reaction mixtures (lanes 4 and 8) contained nonradiolabeled 5′(−) RNA in molar excess. Following incubation, complexes were resolved on a 4% native polyacrylamide gel. The unbound probe is indicated on the left, as are the putative monomeric (L) and multimeric (H) complexes.