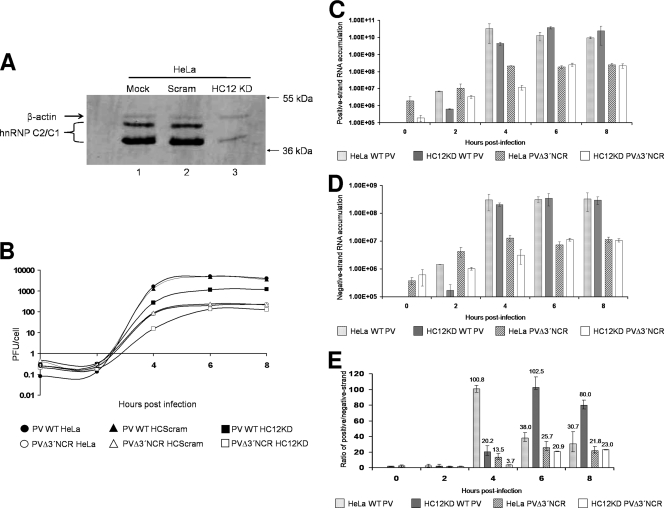
FIG. 8.
Poliovirus growth kinetics and viral-RNA accumulation in hnRNP C-depleted HeLa cells. Knockdown of hnRNP C via retrovirus infection and selection of infected cells is described in Materials and Methods. Plaque assays were carried out as described previously (13) and in Materials and Methods. (A) Whole-cell lysates of mock-treated (lane 1), HCScram-treated (lane 2), and HC12KD-treated (lane 3) cells were generated as described in Materials and Methods. Ten micrograms of each cell lysate was resolved on a 12.5% polyacrylamide-SDS-containing gel. Western blot analysis was carried out with a mouse monoclonal anti-hnRNP C1/C2 antibody (Abcam) and an alkaline phosphatase-conjugated anti-mouse secondary antibody (Promega). As a loading control, mouse monoclonal antibodies recognizing β-actin were utilized (Abcam). (B) One-step growth analysis was carried out in HeLa, HeLaScram, or HeLaHC12KD monolayers in 60-mm plates at 37°C. The cells were infected at an MOI of 25 with wild-type or PVΔ3′NCR virus. Infected HeLa, HeLaScram, and HeLaHC12KD cells were harvested beginning at 0 h postinfection and every 2 h thereafter. The virus particles were released by freeze-thaw, and the total virus yield was determined from the plaque assay titer divided by the total initial cell count. (C) Quantitative real-time PCR of poliovirus positive-strand RNA accumulation was carried out using total-RNA harvests of monolayers from wild-type HeLa cells or retrovirus-infected HeLaHC12KD cells infected with either wild-type poliovirus or PVΔ3′NCR. The cells were infected at an MOI of 25, and total RNA was harvested as described in Materials and Methods. (D) Quantitative real-time PCR of poliovirus negative-strand RNA accumulation was carried out as described for panel C, except that DNA oligonucleotides annealing to poliovirus negative-strand RNA were used. (E) Viral-RNA synthesis is depicted as the ratio of detected positive- to negative-strand RNAs, with the mean ratio depicted above the bars for each sample. As an internal control for RNA content in individual samples, quantitative real-time PCR of cellular β-actin RNAs was carried out for each sample in positive-strand vRNA accumulation (C) and negative-strand vRNA accumulation (D) to normalize poliovirus RNA detection results. Note that for the quantitative real-time PCR assays, shRNA-mediated knockdown was compared to untreated HeLa cells because there were no significant differences in poliovirus growth curves when poliovirus infections of untreated HeLa cells were compared to HeLaScram-treated cells. The error bars indicate standard deviations.