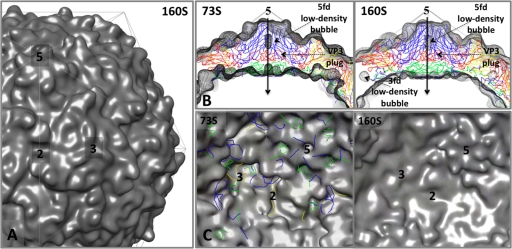
FIG. 4.
Density maps made from atomic models of the native-antigenic empty capsid assembly intermediate (73S) and the mature, native capsid (160S). These two structures are nearly identical on their exterior surfaces but differ dramatically at the interior surface. The transition from 73S to 160S is marked by cleavage of the precursor protein myrVP0 into myrVP4 and VP2 and by encapsidation of viral RNA. The density maps (shown superimposed on an icosahedral framework) were generated by filtering the atomic models from crystal structures (73S from PDB entry 1POV [4]; 160S from PDB entry1HSX [23]) to 10 Å resolution and applying a temperature factor of 300. (A) Outer surface of 160S. (B) Central sections from the density maps are shown in gray mesh, with 73S on the left and 160S on the right, superimposed on a ribbon representation of the 160S atomic model. Certain interior features and the 5-fold-symmetry axes are labeled. (C) The inner surface of the capsid, viewed along the 2-fold-symmetry axis, in 73S (left) and 160S (right). To provide landmarks, ribbon representations of the 160S atomic model are provided, with VP1 in blue, VP2 in yellow, VP3 in red, and myrVP4 in green. Symmetry axes are labeled 2, 3, and 5. On its inner surface, 73S (left) displays an oblong depression at each 2-fold axis and a deep trefoil depression at each 3-fold axis. In 160S (right), these depressions have been filled in by ordered polypeptides: the trefoil depression is filled by the products of myrVP0 cleavage (the carboxyl terminus of myrVP4 and amino terminus of VP2), together with 1028 to 1054 of VP1, and 3160 to 3161 of VP3; the 2-fold depression is filled by polypeptides 1048 to 1067 and 2045 to 2056. Several of these dynamic peptides become rearranged further during cell entry.