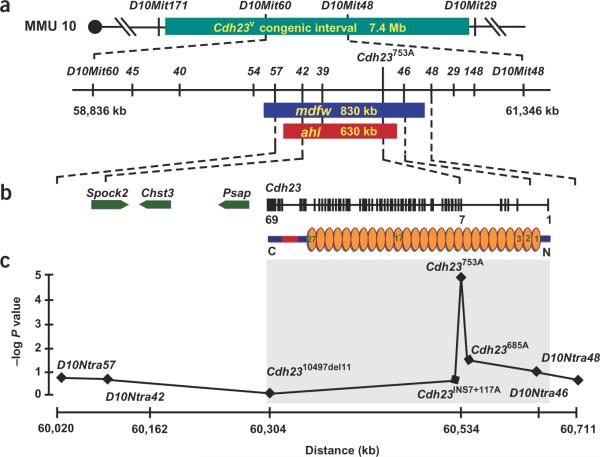
Figure 1.
Positional cloning of ahl and mdfw. (a) Physical map of ahl and mdfw. The Cdh23v congenic interval (7.4 Mb) is defined by D10Mit171 and D10Mit29. Positions of newly developed SNP markers (D10Ntra45, D10Ntra40, D10Ntra54, D10Ntra57, D10Ntra42, D10Ntra39, D10Ntra46, D10Ntra48, D10Ntra29 and D10Ntra148; stems omitted in figure) in relation to mdfw and ahl intervals are shown. D10Ntra54 and D10Ntra48 are recombinant (D10Ntra54 - 0.26 ± 0.18 cM - mdfw - 0.13 ± 0.13 cM - D10Ntra48) with mdfw. The highest lod score for ahl (108.4) was in the region between D10Ntra57 and D10Ntra46. Primer sequences are available on request. (b) Four genes localize to the critical interval: sparc/osteonectin 2 (Spock2), carbohydrate sulfotransferase 3 (Chst3), prosaposin (Psap) and cadherin 23 (Cdh23). Transcription orientation is indicated. Genomic structure, from telomere to centromere, of Cdh23 (black vertical lines) and the domain structure of cadherin 23, including transmembrane domain (red) and ectodomains (orange, 1–27), are shown. (c) Profile of probability scores. Negative logarithm of P is plotted against marker location on the physical map (available at http://genome.cse.ucsc.edu; February 2002 assembly).