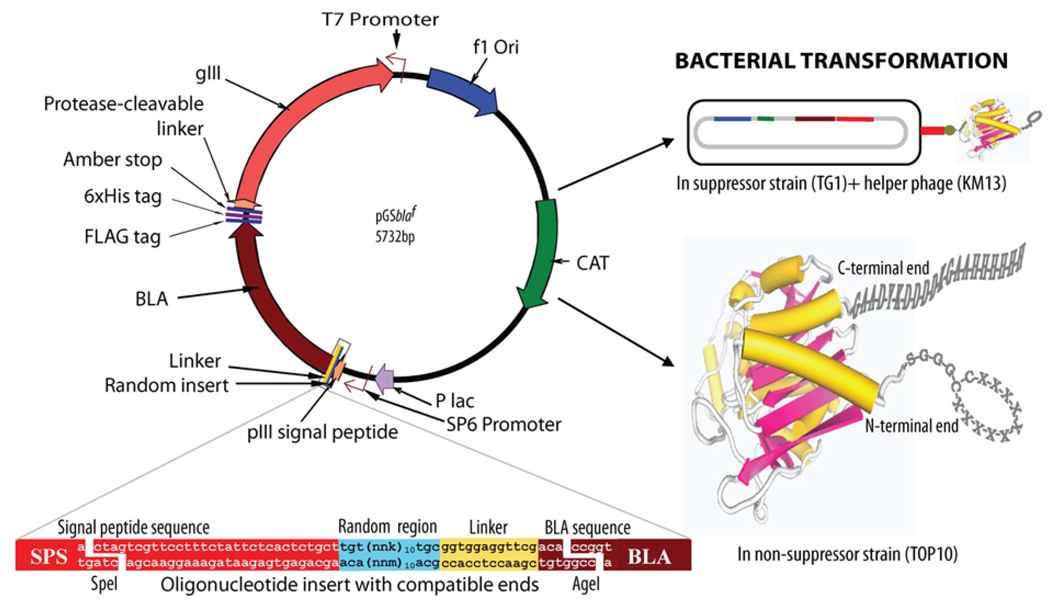
Fig. (1). Schematic presentation of phagemid vector design and features.
The linear dodecapeptide (X12) and cysteine-constrained decapeptide (CX10C) libraries were created at the N-terminal position of Enterobacter cloacae P99 cephalosporinase (BLA) molecule, between the signal peptide and enzyme protein. The random sequences of these libraries were linked to the N-terminal end of the β-lactamase molecule with a short linker (GGGS). Restriction sites SpeI (3326–3231 bp) and AgeI (3256–3261 bp) were used for cloning. The selected site was randomized using the nnk-scheme (n= a, t, c, or g; k=g or t). The N-terminal presence of pIII signal peptide helped in periplasmic translocation of BLA. Chloramphenicol acetyltransferase (CAT) and BLA provided antibiotic resistance to transformed host bacteria in presence of chloramphenicol and cefotaxime, respectively. This vector in suppressor host TG1 E. coli with the help of helper phage (KM13)-generated phage particles expressing BLA-linker-random peptide as an N-terminal fusion protein to phage coat pIII. A protease-cleavable linker between pIII protein of phage particle and BLA was used for trypsin elution of phage following panning. The transformation of non-suppressor strain Top10 E. coli, which recognizes the amber stop (tag) between the BLA and protease-cleavable linker, with the vector allowed the expression of free BLA protein with a random peptide library at the N-terminal end, and FLAG and 6xHis tags at the C-terminal end. The magnified insert region given at the lower left of the figure is inversed in relation to the vector because the insert oligonucleotide and the gene sequences are written 5′– 3′.