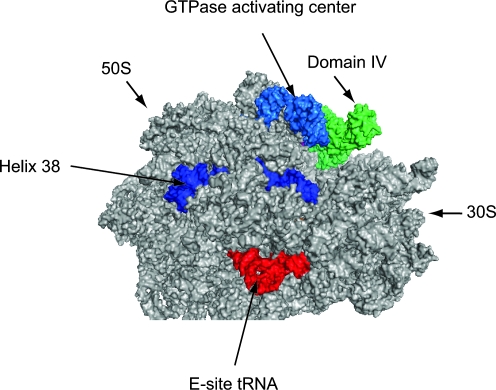
FIG. 3.
Modeling eEFSec on the ribosome. The cryo-EM model of EF-Tu/Phe-tRNA in the ribosomal pre-accommodated state (PDB coordinates 3EP2) was overlayed onto the high resolution crystal structure of the T. thermophilus 70S ribosome in a post-translocation state containing E and P-site tRNAs and an empty A site (PDB coordinates 1VSP[50S] and 2QNH[30S]). Once the pre-accommodated cryoEM structure was aligned, we then overlaid the crystal structure of GDPNP bound archaeal SelB ( PDB coordinates 1W3B; 38) to derive the final high resolution model shown here. Archaeal SelB is shown in green. 50S and 30S ribosomal subunits are indicated. The GTPase activating center (light blue), Helix 38 of the 50S subunit (dark blue), and E-site tRNA (red) are shown as landmarks. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).