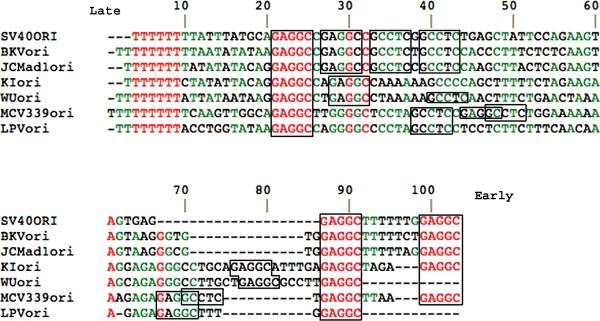
Figure 1. Distinct sequence arrangements in origins of DNA replication of newly discovered human polyomaviruses.
DNA sequences, shown 5’ to 3’ for one strand, were compared for origins of DNA replication of the known human polyomaviruses and for Simian virus 40 (SV40) and Monkey B-cell lymphotropic polyomavirus (LPV) using Multalin [71]. SV40 is included in the figure because of its presence in humans. LPV is included because many humans are reportedly seropositive for it or a homologous virus [65]. Sequences were obtained as described in Box 1. Red lettering denotes 100% identity among all the origins; green lettering denotes >50% identity. Numbering is from the first residue shown for MCV. T-antigen-binding elements, GAGGC or the complement GCCTC, are boxed. MCV is the only virus shown to have the overlapping, palindromic element GAGGCCTC. MCV has two of these elements opposing each other enclosing a highly A-rich segment. These two unusual elements[16] could affect binding of T-antigen hexamers and initiation of DNA replication.