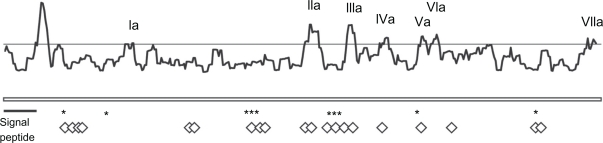
Figure 1.
Reverse conservation analysis of chi18-5 orthologs. Amino acid diversity was estimated using Rate4Site, based on a Clustal X alignment of chi18-5 Trichoderma orthologs, and plotted as W mean scores. The y-axis represents arbitrary units (not shown) while a horizontal line indicates a 0.5 standard deviation cutoff. The x-axis represents residue position, asterisks (*) indicate positions of catalytic residues, diamonds (⋄) indicate substrate-interacting residues. The positions of the signal peptide and regions with high amino acid diversity successfully visualised by homology modelling are indicated (Ia–VIIa).