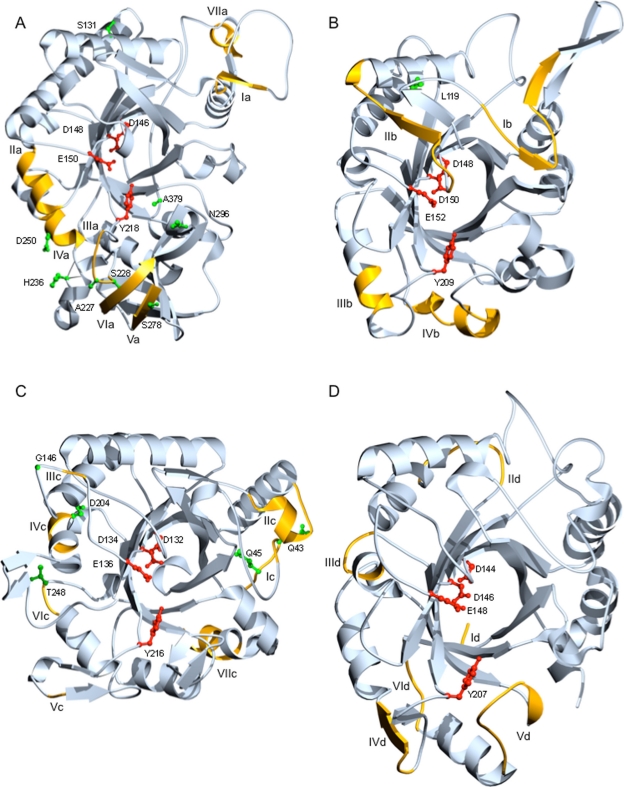
Figure 2.
Homology models of H. jecorina chitinases. Homology models of the catalytic modules of H. jecorina chitinases (A) chi18-5, (B) chi18-13, (C) chi18-15 and (D) chi18-17 were generated using SOD and adjusted in O, based on hidden Markov models and Clustal W amino acid sequence alignments. Conserved catalytically important residues are indicated in red, amino acids under strong positive selection (Bayes factor ≥50) are indicated in green, variable regions from reverse conservation analysis (I scores ≥0.5) are indicated in orange and marked in Roman numerals from N- to C-termini. Residue numbering refers to catalytic module sequences used for modelling; 15–424 (chi18-5), 30–320 (chi18-13), 25–322 (chi18-15), 28–311 (chi18-17).