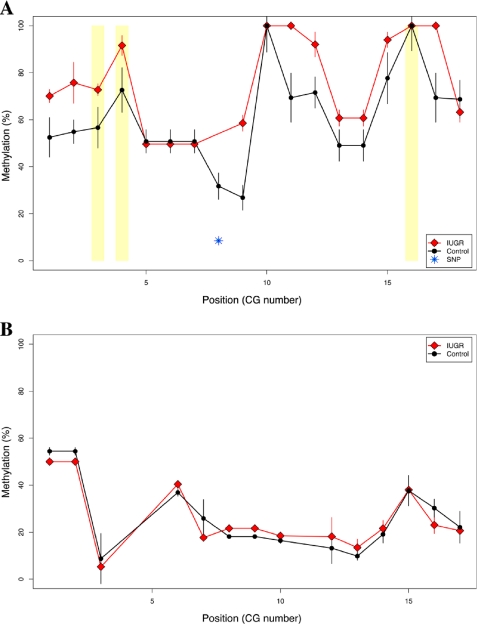
FIGURE 3.
Bisulfite MassArray validation confirms IUGR-specific hypermethylation of the Gch1 locus. MassArray was performed for two genomic regions as described by the PCR conditions in supplemental Table 1. Three replicate assays were performed for each of three IUGR offspring and controls. A, MassArray validation results are shown as group-median percent methylation values with standard error bars (y axis) for each individual CG site (in sequential order along the x axis). IUGR is depicted with red lines and diamonds, whereas control data are shown in black lines and filled circles. HpaII sites are highlighted by yellow rectangles. One single nucleotide polymorphism (SNP) was identified in the underlying genomic sequence and was removed from affected samples in the dataset (blue, starred symbol). B, MassArray data are similarly shown for a second genomic region at the Pdx1 gene. No differences in DNA methylation were observed at this age, with relative hypomethylation in all samples.