FIGURE 1.
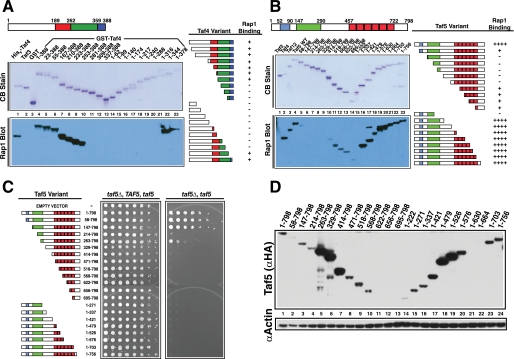
Mapping the RBDs of Taf4 and Taf5. A, schematic of Taf4 nonconserved (white) and conserved residues (colored). N- and C-terminal truncations were expressed, purified, and fractionated by SDS-PAGE and blotted; one blot was stained with Coomassie Blue (CB Stain), and the other was probed with Rap1; Taf4-bound Rap1 was detected with anti-Rap1 IgG (Rap1 Blot). Lane 1 contained positive control His6-Taf4; lanes 2 and 3 contained non-Rap1-binding negative controls, His6-Taf3 and glutathione S-transferase (GST). Taf4 amino acids fused to glutathione S-transferase are indicated above the Coomassie Blue-stained blot, lanes 4–23. Summary of Rap1-Taf4 binding data (right). B, schematic of Taf5 showing nonconserved (white) and conserved sequence elements (colored). N- or C-terminally truncated forms of Taf5 (lanes 4–23) were generated, purified, and tested as in A. Lane 1 contained the negative control, non-Rap1-binding Taf3, and lanes 2 and 3 contained positive control proteins His6-Taf4 and His6-Taf12. Summary of Rap1-Taf5 binding data (right). C, plasmid shuffle complementation assay testing the ability of various truncated forms of Taf5 to support viability. Serial 1:4 dilutions of cells expressing the indicated Taf5 variants were subjected to growth as pseudodiploids (relevant genetic constitution: taf5Δ, TAF5, taf5) or following loss of the URA3-marked TAF5 covering plasmid, scored as 5-fluoroorotic acid resistance (taf5Δ, taf5). Plates were incubated for 3 days at 30 °C. D, steady state protein levels of WT and Taf5 variants measured in whole cell extracts by immunoblotting with anti-HA (Taf5 (αHA)) or anti-actin (αActin) IgGs; actin served as extraction/loading control.