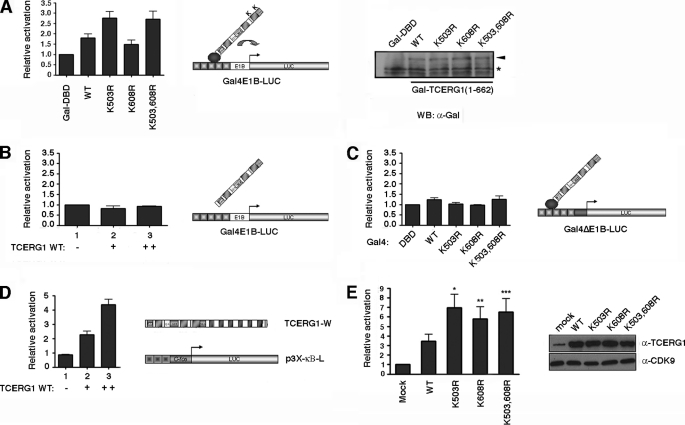
FIGURE 5.
SUMO modification sites negatively regulate the TCERG1-mediated transcriptional activation. A, HEK293T cells were cotransfected with a Gal4-luciferase reporter plasmid (Gal4E1B-LUC), a control plasmid expressing the Gal4 DNA binding domain (Gal4-DBD, 1st lane), the wild-type amino-terminal domain of TCERG1(134–662) (lane 2), or the SUMO mutant variants (K503R, 3rd lane; K608R, 4th lane; K503R,K608R (K503,608R), lane 5) fused to the Gal4 DNA-binding domain. The luciferase activity is shown relative to the Gal4-DBD that was set at 1. The data shown are from four independent experiments performed in triplicate (p (wild type (WT) versus Gal4-DBD) = 0.02; p (wild type versus K503R) = 0.0058; p (wild type versus K608R) = 0.11; p (wild type versus K503R,K608R) = 0.01; paired Student's t test, set at p < 0.05). A sample of cell lysate was immunoblotted with anti-Gal4 antibody to demonstrate the expression levels. B, HEK293T cells were cotransfected with a Gal4-luciferase reporter plasmid (Gal4E1B-LUC) and a control plasmid (lane 1) or increasing amounts of TCERG1(134–662) without the Gal4 DNA-binding domain (lanes 2 and 3). The luciferase activity is shown relative to the control that was set at 1. The data shown are from four independent experiments performed in triplicate. C, HEK293T cells were cotransfected with a Gal4-luciferase reporter plasmid lacking a functional TATA-box (Gal4ΔE1B-LUC), a control plasmid expressing the Gal4 DNA binding domain (Gal4-DBD, 1st lane), the wild-type amino-terminal domain of TCERG1(134–662) (2nd lane) or the SUMO mutant variants (K503R, 3rd lane; K608R, 4th lane; K503,608R, 5th lane) fused to the Gal4 DNA-binding domain. The luciferase activity is shown relative to the Gal4-DBD that was set at 1. The data shown are from four independent experiments performed in triplicate. D, HEK293T cells were cotransfected with the p3X-kb-L reporter plasmid together with increasing concentrations of wild-type TCERG1 expression plasmid. The luciferase activity was calculated relative to the control that was cotransfected with the reporter and empty plasmids. The total DNA amount for transfection was kept the same in each sample by normalizing with empty vector. E, HEK293T cells were cotransfected with the p3X-kb-L reporter plasmid together with empty vector (1st lane), wild-type TCERG1 (2nd lane), and SUMO variants (K503R, 3rd lane; K608R, 4th lane; K503,608R, 5th lane) DNA constructs. The luciferase activity was calculated relative to the control. The data shown are from four independent experiments performed in triplicate (*, p < 0.01; **, p < 0.02; ***, p < 0.04). Cell lysates were analyzed by immunoblotting with the indicated antibodies to detect the TCERG1 and CDK9 proteins.