Abstract
We present the results of combining design and selection to remodel a protein–peptide binding interface, using the peptide PTIEEVD and the TPR1 module interaction as our test case. We initially used the program Rosetta to interrogate possible TPR1 sequences compatible with binding the peptide PTIEEVD. Based on these results, we screened a small library of TPR1 variants, using a split GFP fluorescent assay to identify proteins that are able to bind to the PTIEEVD peptide. We discuss the similarities and differences between the modeling and selection results at each position. We show that a new ‘consensus’ TPR1, created based on the results of the sequences identified in the screen, indeed binds to the PTIEEVD peptide. These results demonstrate the utility of combining design and selection in a synergistic fashion to remodel protein recognition interfaces.
Keywords: peptide binding, protein design, Rosetta Design, split-GFP assay, tetratricopeptide repeat protein 1 (TPR1)
Introduction
An important goal of protein engineering is to create proteins with novel-binding activities. Such proteins would have widespread practical applications; for example, in molecular and cellular biology they could replace antibodies for affinity purifications and cellular localization studies; in analytical and medicinal chemistry they could be used as bio-sensors to select specific compounds from complex mixtures or could be used to identify and target particular cells within an organism (Binz et al., 2004; Cortajarena et al., 2008; Skerra, 2008; Jackrel et al., 2009). Finally, redesigning protein–peptide interactions and testing the results is important, because it deepens our fundamental understanding of the underlying physical chemistry of molecular recognition (Cortajarena et al., 2004).
There are two extreme approaches to creating a protein with a novel binding activity—‘rational’ structure-based design or random mutagenesis in combination with a screen or selection for the desired activity (Binz and Pluckthun, 2005; Binz et al., 2005; Jiang et al., 2008; Rothlisberger et al., 2008; Jackrel et al., 2009; Karanicolas and Kuhlman, 2009; Mandell and Kortemme, 2009; Thyme et al., 2009). Here we report how design and selection can be combined to synergistic effect. We chose a well-defined protein—peptide system for these test studies, specifically the tetratricopeptide repeat 1 (TPR1) domain of HOP and its cognate ligand, the C-terminal amino acids of Hsp70 (PTIEEVD) (Scheufler et al., 2000; Yi et al., 2009). Our goal was to identify different combinations of residues in the TPR-binding cleft that are compatible with binding this peptide. We first used a computer-based search (Das and Baker, 2008) to identify potential substitutions at the protein–ligand interface that are well suited for ligand binding. After several rounds of such modeling, we identified several positions where alternative amino acids that are compatible with binding the PTIEEVD peptide are predicted. Guided by these results, we created a targeted library in which each potentially substitutable position is allowed to vary within parameters derived from the computer-based design; for example small and hydrophobic, or charged, or large and aromatic.
We subsequently created and screened the library, by fluorescence assay on agar plates, to identify clones that expressed a TPR variant that binds to PTIEEVD. We then made a consensus protein from the sequence of positive clones and tested its binding. We found that at some positions functional substitutions can be well predicted by computational design, while at other positions a screen allows us to better chose between apparently equally good alternatives. At a third class of positions, the screen reveals a preference for amino acids that were not the top choice from the modeling stages. Thus we are presenting a general protein design methodology that advantageously combines computer-based design with in vivo selection to create proteins with novel-binding sites.
Materials and methods
Computational methods
TPR1 + Hsp70 peptide coordinates were obtained from the Protein Data Bank (PDB ref. code 1ELW) (Scheufler et al., 2000). Deep View v4.0 Swiss PDB viewer was used to minimize the structure using GROMOS 6 in 100 steps. The lowest energy conformation was exported to Rosetta Design and hyper-variable residues 2, 5, 9, 12, 13, 33, 34, where numbering corresponds to the position within one TPR motif, were matched with the numbering in TPR1 + His70 co-crystal structure. These seven residues appear in each of the three TPR motifs resulting in 21 hyper-variable residues within 1ELW. Residue 1 of TPR1 corresponds to residue 4 of HOP.
To find the lowest energy conformation, 21 hypervariable residues were then selected and allowed to vary to all amino acids. After this first round of calculation we selected all residues that changed from the wild-type residue into non-Ala amino acid—a total of 11 positions. Further, we ran another 100 rounds to determine the sustained variability of the selected 11 residues. Next, we measured distances between residues 5, 8, 12, 16, 36, 37, 46, 50, 58, 73 and 83 and the peptide in an energy-minimized structure. We have selected residues within 4 Å (8, 46, 50, 73 and 83) from the peptide for further calculations, assuming that these residues will make the major contribution to the binding energy.
The positions 8, 46, 50, 73 and 83 were then allowed to vary in Rosetta to all amino acids and all allowed rotamers. We initiated the calculations 20 times (20 parallel runs) to determine the frequency of occurrence of certain amino acids at a particular position.
Cloning
All enzymes for cloning were purchased from New England Biolabs (Beverly, MA), unless otherwise noted. All oligonucleotide synthesis and sequencing was performed by the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University.
To assemble the TPR1 library, the TPR1 template was synthesized from six overlapping nucleotides: (1) 5′-agcaggtcaatgagctgMRggagaaaggcaacaa ggccctgagcgtgggtaacatcgatgatgccttacagtgctactcc-3′; (2) 5′-cttctggtagtctcc tttNRNggcataggcgWRagaacggttgctatacagcacgtggttgtggggatccagcttaatagcttcggagtagcactgtaaggc-3′, (3) 5′aaaggagactaccagaaggcttatgaggatggctgcaagactgt cgacctaaagcctgactggggcMRaggctattcacgaaaagcag-3′; (4) 5′-gcttggcttcttcaaag cggtttaagaatcgtagagctgctgcttttcgtgaatagcc-3′; (5) 5′-gctttgaagaagccaagcga acctatgaggagggcttaaaacacgaggcaaataatcctcaactgaaagag-3′; and (6) 5′-cctggcctcc atattctgtaaaccctctttcagttgaggatt-3′. At each position of randomization an equimolar mixture of specific bases was added, where M denotes A, C; R denotes A, G; W denotes A, T and N denotes A, G, C, T. Four sites were randomized as explained in the main text. Oligonucleotides 1 and 2, 3 and 4, 5 and 6 were joined by Klenow extension, and this was followed by a series of two PCR amplifications. The first amplification fused oligonucleotides 3–6 using the primers 5′-aaaggagactaccagaag-3′ and 5′-attattgacgtccccctggcctccatattctg-3′. A final PCR amplification joined two remaining fragments using the primers 5′-taataaccatggagcaggtcaaatgagctg-3′ and 5′-attattgacgtccccctggcctccatattctg-3′. The library of inserts was then double digested with AatII and NcoI and ligated into the C-teminal part of split-GFP.
The synthetic gene encoding for TPR1 C7 was created by the same strategy as the library using six overlapping nucleotides: (1) 5′-agcaggtcaatgagctgagggagaaaggcaacaaggccctgagcgtgggtaacatcgatgatgccttacagtgctactcc-3′; (2) 5′-cttctggtagtctcctttaaaggcataggcgagagaacggttgctataca gcacgtggttgtggggatccagcttaatagcttcggagtagcactgtaaggc-3′; (3) 5′-aaaggaga ctaccagaaggcttatgaggatggctgcaagactgtcgacctaaagcctgactggggccgaggctattcacgaaaagcag-3′; (4) 5′-gcttggcttcttcaaagcggtttaagaatcgtagagctgctgcttttcgt gaatagcc-3′; (5) 5′-gctttgaagaagccaagcgaacctatgaggagggcttaaaacacgaggcaaat aatcctcaactgaaagag-3′; (6) 5′-cctggcctccatattctgtaaaccctctttcagttgaggatt-3′. The primers for two rounds of PCR amplifications were: 5′-aaaggagactaccagaag-3′ and 5′-aataatttcgaatcacctggcctccatattctg-3′, for fusing oligonucleotides 3–6, and 5′-taataaggatccagcaggtcaatgagctg-3′ and 5′-aataatttcgaatcacctggcctccatattctg-3′ for final joining of fragments. The nucleotide was then double digested with BamHI and HindIII and ligated into pProEx-HTA vector (GibcoBRL, Gaithesburg, MD) to create a gene with an N-terminal His6-tag followed by TEV cleavage site. The final construct was sequenced to verify its identity.
Protein expression and purification
The plasmids were transformed into E.coli BL21. Overnight cultures were diluted 1:100 in 1 l of Luria broth at 37°C, with shaking at 250 rpm, and were grown to an OD600 of 0.5–0.8. Expression was induced with 1 mM IPTG, followed by overnight incubation at 18°C. The cells were harvested at 7000g for 30 min and the pellets were frozen at −20°C until purification.
To purify the TPR proteins, the pellets were thawed by resuspending in lysis buffer (50 mM Tris, 300 mM NaCl, 5 mM β-mercaptoethanol, 10% (v/v) glycerol, 0.1% (v/v) Triton X) with one tablet of complete EDTA-free protease inhibitor cocktail and 400 mg of lysozyme. The suspension was sonicated, and the lysate cleared by centrifugation for 1 h at 17 000g. The proteins were then purified using an Ni-NTA resin (Qiagen) according to the manufacturer's protocols.
The proteins were further purified with size-exclusion chromatography using S-200 16/60 HR column (Amersham Pharmacia). The protein-containing fractions were collected, concentrated and dialyzed into phosphate buffer (25 mM Na2HPO4, pH 7.4, 50 mM NaCl) supplemented with 5 mM DTT. All protein concentrations were determined by measuring UV absorption at 280 nm using extinction coefficients calculated from amino acid composition. All experiments were carried out using His-tagged proteins.
Circular dichroism measurements
Circular dichroism spectra were acquired using 10 μM protein samples in phosphate buffer using an Applied Photophysics Chirascan CD spectrophotometer (Applied Photophysics, Leatherhead, Surrey, UK).
Far-UV CD (190–260 nm) spectra were recorded at 25°C to assess the secondary structure of the TPRs. Thermal denaturation curves were recorded by monitoring molar ellipticity at 222 nm while heating from 4 to 94°C in 1°C increments with an equilibration time of 5 min at each temperature. We do not report Tm or ΔG because these are not reversible.
Fluorescence anisotropy
To determine the binding affinities, increasing amounts of protein, TPR1 WT or TPR1 C7, were titrated to an N-terminal fluorescein-labeled C-terminal 10-mer peptide of Hsp70 in 25 mM Na2HPO4 pH 7.4, 50 mM NaCl, 5 mM DTT buffer. Binding was performed at 50 nM peptide concentration in a 0.2 cm path-length cuvette at 25°C, and the fluorescence anisotropy was recorded after 5 min equilibration. Fluorescence anisotropy experiments were recorded in a PTI Quantamaster C-61 two-channel fluorescence spectrophotometer equipped with excitation and emission polarizers. Excitation was achieved with a 6 nm slit-width at 492 nm and the emission recorded at 516 nm with slit-width of 6 nm.
For excitation at the vertical orientation (0°) the anisotropy (r) is:
where G is the G-factor, IVV and IVH are the vertical and horizontal emission of the sample, respectively, and IB,VV and IB,VH are the intensity of the emission of the blank with emission polarizer at vertical and horizontal orientation, respectively. The G-factor corrections were calculated using the equation: G = (IHV – IB,HV)/(IHH – IB,HH), where IHV is the vertical emission (0°) of a standard solution with excitation in horizontal orientation (90°), IHH is the horizontal emission (90°) of a standard solution with excitation in vertical orientation (0°), IB,HV is the vertical emission (0°) of a blank solution with excitation in horizontal orientation (90°) and IB,HH is the horizontal emission (90°) of a blank solution with excitation in vertical orientation (0°) using phosphate buffer as a blank solution and a 50 nM fluorescein-labeled C-terminal 10-mer peptide of Hsp70 as a standard solution.
The fraction of peptide bound at each point in the binding curve was calculated by the equation:
where r is the observed anisotropy of the peptide at any protein concentration, rf is the anisotropy of the free peptide and rb is the anisotropy of the peptide in the plateau region of the binding curve. The data were fit to a single-site binding model using SigmaPlot 8 (Systat Software, Point Richmond, CA, USA)
where KD and [P] are the dissociation constant and protein concentration, respectively.
Results
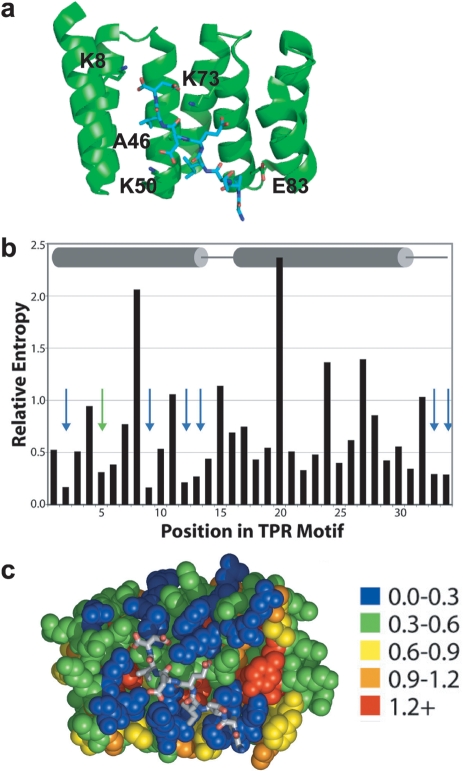
The TPR is a 34 amino acid helix-turn-helix motif (helices A1 and A2), which occurs in many proteins, and functions as a protein–protein interaction module (Lamb et al., 1995; D'Andrea and Regan, 2003; Kajander et al., 2005; Cortajarena and Regan, 2006). The binding characteristics of several natural TPR-peptide pairs have been well characterized and high-resolution crystal structures are available (Das et al., 1998; Scheufler et al., 2000; Kajander et al., 2009). Typically, the peptide ligand is bound in an extended conformation. An attractive feature of TPR-peptide recognition, especially for computational design, is that there is no evidence for any substantial changes in the backbone of the TPR associated with peptide binding (Grove et al., 2008). Here, we report the redesign of the ligand-binding interface of the TPR1 domain of Hsp Organizing Protein (HOP) with its cognate ligand, the C-terminal amino acids of Hsp70. Figure 1A shows the co-crystal structure of TPR1 in complex with a peptide corresponding to the seven C-terminal amino acids of Hsp70 (PTIEEVD). The PTIEEVD peptide is bound in an extended conformation with the side chains of Pro, Ile, Val and Asp, along with the C-terminal carboxylate in contact with the side chains of residues of TPR1 in the binding cleft. Additional TPR side-chain to peptide main-chain interactions are also present.
Fig. 1.
Ligand binding by TPR1 domain. (a) Co-crystal structure of TPR1 with its ligand, C-terminal peptide of Hsp70 (PDB ID: 1elw) (Scheufler et al., 2000). (b) The relative entropy values are shown for each TPR position, with secondary structure indicated (cylinders represent helices and lines represent loops). Arrows indicate the positions of the seven most variable residues. (c) Relative entropy values (color coding is shown) are mapped onto the co-crystal structures of HOP-TPR1/Hsp70 peptide. Adapted from (Magliery and Regan, 2005).
Rosetta Design
Our goal was to identify mutations in the binding site of TPR1 that are compatible with cognate-ligand binding. In other words, we sought to find different TPR1 sequences that can bind the same peptide ligand. First, we considered all hypervariable residues, i.e. positions not conserved among TPRs, on TPR1. We have previously described seven positions that are hypervariable when the amino acid sequences of all TPRs are aligned (Fig. 1B) (Magliery and Regan, 2005). Positions 2, 5, 9, 12, 13, 33 and 34 are the residues most likely involved in ligand recognition within a TPR repeat (Fig. 1B and C). Not every TPR repeat in a given binding module necessarily uses all these residues for ligand recognition. In Fig. 1C, residues in TPR1 are color-coded based on their variability/conservation with the most hypervariable ones, 18 in the three TPR repeats of TPR1, colored blue. It is evident that most hypervariable positions correspond to surface-exposed residues in the ligand-binding site.
We used the program Rosetta (Das and Baker, 2008) to assess the alternative TPR1-peptide interfaces. We started with the energy-minimized structure of the TPR1-PTIEEVD complex. We kept all the residues other than the 21 hypervariable residues fixed in their starting identity and position, and allowed only the hypervariable residues to vary.
In the first round of modeling (Table I), we found that 11 of the 21 hypervariable positions were consistently changed from the wild-type residue into a non-Ala amino acid. We also observed a tendency for Rosetta to make changes into Ala, presumably to avoid steric clashes by removing unfavorable packing energy, but without adding favorable interaction energy. We disregarded this class of substitutions. All other positions were unchanged.
Table I.
Results of Rosetta modeling where 21 hypervariable residues were allowed to vary to any of the 20 amino acids
| Residue # in TPR Repeat | Residue # in TPR1 | WT residue | Rosetta prediction |
|---|---|---|---|
| 2 | 5 | Asn | Asp |
| 5 | 8 | Lys | Arg |
| 9 | 12 | Asn | Asp |
| 12 | 15 | Leu | / |
| 13 | 16 | Ser | Lys |
| 33 | 36 | His | Thr |
| 34 | 37 | Asn | Arg |
| 2 | 43 | Asn | / |
| 5 | 46 | Ala | / |
| 9 | 50 | Lys | Leu |
| 12 | 53 | Asp | / |
| 13 | 54 | Tyr | Phe |
| 33 | 74 | Gly | / |
| 34 | 75 | Tyr | / |
| 2 | 73 | Lys | Arg |
| 5 | 76 | Ser | Ala |
| 9 | 80 | Ala | / |
| 12 | 83 | Glu | Arg |
| 13 | 84 | Phe | / |
| 33 | 104 | Asn | / |
| 34 | 105 | Asn | / |
In the next round of modeling, all 11 potentially mutatable residues that changed in the first round of modeling were allowed to vary in both identity and in side-chain rotamers. We found that the changes were consistent between two rounds of Rosetta modeling and the results therefore independent of in silico library size (21 vs. 11 positions mutated). The remaining 10 hypervariable positions were kept as in the original energy-minimized structure.
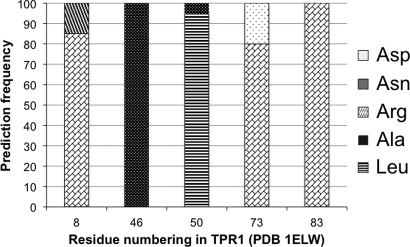
Of the 11 variable residues identified in the second round of modeling, we chose five residues within 4 Å of the peptide for more extensive modeling trials (Table II). Although 4 Å is a somewhat arbitrary cut-off, we made the assumption that residues closest to the peptide will have the largest effect on binding. All residues were kept as in wild type except for the five residues in the binding pocket, close to the peptide. These five residues were allowed to change into any amino acid, with any rotamer. We repeated this last round of Rosetta modeling 20 times and the frequencies of amino acid occurrences at each position are shown in Fig. 2. It is interesting to note that although we did not obtain the same sequence in every one of 20 parallel rounds of modeling, the sequences coalesce around a unique solution. Positions 46 and 83 remained unchanged over 20 rounds of modeling, whereas positions 8, 50 and 73 were somewhat variable between different rounds.
Table II.
Final Rosetta Design predictions, combinatorial library and consensus sequence
| Position | WT | Rosetta prediction | Library | Codon | Consensus |
|---|---|---|---|---|---|
| 8 | K | R | K/R/Q | MRG | R |
| 46 | A | A | Y/F/H/L | RWC | L |
| 50 | K | L | F/S/L/P/I/T/M/V/A | NRN | F |
| 73 | K | R | K/R/Q | MRG | R |
| 83 | E | R | R | CGA | R |
Fig. 2.
Prediction frequency of Rosetta Design. In the 20 independent rounds of modeling of five amino acids inside the peptide-binding pocket, sequences coalesce around unique solution shown in the third column of Table II.
Guided by modeling results, we constructed a small combinatorial library in which we randomized four of five residues previously discussed (shown in Table II) and restricted the identities of amino acids in those positions. In positions 8 and 73 we allowed wild-type Lys to vary between Lys, Arg and Gln. Lys 50 was allowed to vary to small hydrophobic and polar residues, and also to medium-sized amino acids since Rosetta predominantly mutated this position into Leu. We also allowed Ala46, although not mutated in Rosetta, to vary among several hydrophobic amino acids because this residue, together with position 50, forms a hydrophobic cavity for binding of peptide Ile. We hypothesized that a more hydrophobic cavity may increase the affinity of the protein to the hydrophobic part of the peptide.
Targeted mutagenesis plus selection
Rather than synthesizing the gene for each possible TPR1 variant, and testing each purified protein individually, we took advantage of a fluorescent colony assay that allowed us to screen a small TPR1 library on agar plates (Magliery et al., 2005). The composition of the library was based on the Rosetta predictions, and the possible amino acids encoded at each position are summarized in Table II. The theoretical size of this library is 324 protein sequences, encoded by 2048 DNA sequences.
To implement the functional screen, the TPR1 library was fused to the C-terminal half of GFP, and the C-terminal Hsp70 peptide was fused to the N-terminal half of GFP. We have previously shown that only cognate TPR-peptide binding pairs are able to assemble the split GPF and give rise to fluorescent colonies (Magliery et al., 2005, Jackerel et al., 2009).
Cells containing the N-terminal half of GFP fused to the Hsp70 peptide were transformed with the C-terminal half of GFP fused to the TPR1 library, and the cells were plated and incubated to develop fluorescence. Colonies that appeared brightest under UV illumination were chosen, and the TPR1 proteins they encoded were sequenced. The results of sequencing the 40 unique TPR1 clones from bright colonies identified in this fashion are presented in Table III. Because certain amino acids are encoded by multiple codons, the probability of their occurrence is skewed higher (codon bias). We, therefore, normalized the sequencing results for such non-equal codon representation in the library. The last column in Table III shows the amino acid preferences at each position, normalized for library bias.
Table III.
Sequencing results and analysis of codon bias
| Residue number | Amino acid | Occurrence | Bias | Normalized |
|---|---|---|---|---|
| 8 | K | 12 | 1 | 12 |
| R | 24 | 2 | 12 | |
| Q | 4 | 1 | 4 | |
| 46 | Y | 6 | 1 | 6 |
| F | 8 | 1 | 8 | |
| H | 7 | 1 | 7 | |
| L | 19 | 1 | 19 | |
| 50 | F | 10 | 1 | 10 |
| S | 8 | 2 | 4 | |
| L | 6 | 3 | 2 | |
| P | 10 | 2 | 5 | |
| I | 0 | 1.5 | 0 | |
| T | 1 | 2 | 0.5 | |
| M | 1 | 0.5 | 2 | |
| V | 1 | 2 | 0.5 | |
| A | 3 | 2 | 1.5 | |
| 73 | K | 5 | 1 | 5 |
| R | 30 | 2 | 15 | |
| Q | 5 | 1 | 5 | |
| 83 | R | 40 | 1 | 40 |
The residue with highest occurrence is highlighted in bold.
The screen revealed preferences for particular amino acids at each position. At some positions the favored residue matches well with the Rosetta prediction, for example position 8 is Lys in the wild-type TPR1. Rosetta predicts Arg, and experimentally we find Lys and Arg with equal likelihood. At other positions, the residue predicted by Rosetta is not the one found experimentally. The most dramatic example of such behavior is seen at Lys50, where Rosetta predicted Leu, but experimentally Phe was favored.
Analysis of the results summarized in Table III allowed us to derive a ‘consensus’ sequence for all the mutated binding-site positions (last column in Table II). We created a protein that has the wild-type amino acids of TPR1 at all positions except the five binding-site residues, where we substituted the most favored residues at each position from Table III. We named this protein TPR1C7.
TPR1C7 characterization
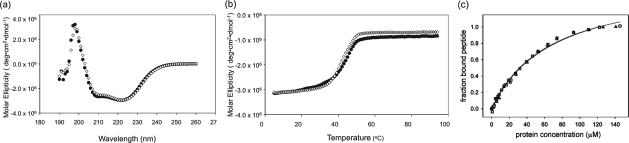
We created the gene encoding TPR1C7 and expressed and purified the protein. By CD, TPR1C7 exhibits a characteristic α-helical signature (Fig. 3A), with minima at 220 and 208 nm, which is super-imposable with the spectrum of wild-type TPR1. The thermal denaturation curves of TPR1 and TRR1C7 are also super-imposable (Fig. 3B). These results were expected, because we had focused on optimizing the TPR1-peptide interface, not on changing the intrinsic structure or stability of TPR1.
Fig. 3.
Characterization of consensus sequence protein C7. (a) CD wavelength scan. (b) Thermal melt followed by decrease of the CD signal at 222 nm. (c) Fluorescence anisotropy of fluorescently labeled Hsp 70 peptide. WT TPR1 is shown as squares and TPR1 C7 as triangles. Note overlapping experimental data in all three panels.
We used fluorescence anisotropy to characterize the interaction of TPR1C7 with the Hsp70 C-terminal peptide. The dissociation constant for the TPR-PTIEEVD peptide interaction is 65 ± 4 μM for wild-type TPR1 (consistent with previously determined values in our laboratory) and 70 ± 7 μM for TPR1C7. Representative binding data are shown in Fig. 3C. It is interesting to note that the TPR scaffold allows for two different sequences in the binding site that have the same affinity for the same peptide.
Discussion
Here we present the results of experiments that combine computer design and genetic selection to remodel the binding interface of a peptide-binding module. Our results highlight the synergy of these two strategies. At some positions, the Rosetta prediction and the results of the guided randomization and selection were completely congruent, for example at positions 8 and 73. At other positions, the selection clearly revealed a preference for an amino acid other than that favored in the Rosetta prediction—for example position 50.
Positions 33 and 34 in the TPR repeat are predicted (from sequence variability) to be involved in ligand binding, but experimental confirmation is lacking (Magliery and Regan, 2005). Rosetta identifies these two positions as variable only for repeat 1 of TPR1 (Table I). Inspection of the co-crystal structure confirms that positions 33 and 34 are in the vicinity of the peptide C-terminus only in repeat 1. These two positions, therefore, offer attractive possibilities for the designs of novel proteins based on the TPR1 scaffold that bind peptides with C-termini longer than that of PTIEEVD.
Another interesting result of Rosetta modeling is charge reversal in certain positions. We experimentally explored mutation E83R. E83 was shown, by electrostatic calculations, to have unfavorable contribution to peptide-binding energy (+1.4 kcal/mol) (Kajander et al., 2009). A previously reported E83Q substitution did not significantly affect the dissociation constant (from 50 to 35 µM), but it is a ‘neutral’ mutation in comparison to the charge reversal that we identified by Rosetta. Lys 73, which is the so-called carboxylate clamp residue, is essential for peptide binding (Scheufler et al., 2000; Cortajarena et al., 2004). It has been recently shown that interaction of Lys 73 with the C-terminal aspartate side chain contribute a smaller energy of stabilization (−0.14 kcal/mol) to the peptide binding than other carboxylate clamp residues such as Lys 8, Arg12, Arg43 (−0.90 kcal/mol) (Kajander et al., 2009). Interestingly, only position 43 of the di-carboxylate clamp remains unchanged in Rosetta modeling (Table I). Therefore, it seems that Rosetta identified previously recognized ‘weak spots’ in the electrostatic interactions between TPR1 and its cognate ligand.
Changes to the residues that contribute to the electrostatic interaction between TPR1 and peptide, such as positions 8 and 73, are conservative (Table II). The most significant changes are in the part of the protein that binds the N-terminal part of the peptide. TPR1 binds to the EEVD peptide with a Kd of ∼300 μM, compared with a Kd of 50 µM for binding to the PTIEEVD peptide. The surface area of peptide buried upon complex formation for PTIEEVD is 1330 Å2 vs. only 650 Å2 for EEVD (Scheufler et al., 2000).
Contacts responsible for the over 20-fold difference in affinity between the two peptides involve hydrophobic and van der Waals interactions of TPR1 with Ile and Pro of the peptide. Ile interacts with TPR1 residues Ala 46, Ala 49, Lys 50 of helix A2, and Pro interacts with TPR1 residues Glu83 and Phe84 of helix A3. Interestingly, although the positions 46, 50, 83 and 84 are ‘hypervariable’, position 49 (corresponding to position 7 in the second TPR repeat) is also a key structurally conserved residue (Scheufler et al., 2000; Magliery and Regan, 2005). Moreover, this is one of the TPR ‘signature’ residues (Magliery and Regan, 2004) that define TPR structure. Therefore, we chose not to change Ala49. Position 84 is the only position that remained unchanged in our initial Rosetta modeling (Table I). Lys 50 was consistently changed to Leu by Rosetta through all rounds of modeling. In comparison to Lys, Leu has the same number of methyl groups but in a branched arrangement, and is uncharged. Phe was experimentally selected in the combinatorial library, and has a larger volume (and hydrophobic surface area) than either Lys or Leu. Although position 46 (Ala) remained consistently unchanged in Rosetta modeling, we mutated this residue to a subset of larger hydrophobic residues. The library screen identified Leu as a consensus residue (Table III). Rosetta consistently changed E83 to Arg, and we made that direct substitution. Arg not only has the opposite charge from Glu, but also a significantly larger volume and area (180 Å3 vs. 143.8 Å3, and 225 Å2 vs. 173 Å2, respectively). All these mutations should theoretically increase hydrophobic interaction between the consensus sequence protein and Ile and Pro of the peptide.
Considering previously discussed changes, it is interesting that affinity of TPR1 C7 for PTIEEVD did not improve over WT TPR1. One possible explanation is that the ‘first coordination sphere’, i.e. residues closest to the peptide, already has optimized interaction energy. Previous work from our laboratory showed that grafting only six binding-site residues on to the consensus TPR scaffold is enough to impart ligand affinity and specificity (Cortajarena et al., 2010). Moreover, varying residues that are not in direct contact with the peptide can modulate this novel affinity (Cortajarena et al., 2004). Thus, to design a TPR1 protein with a higher affinity for the Hsp70 peptide we should perhaps consider the remaining 6 residues of 11 that were mutated in the first round of Rosetta modeling (Table I). Nevertheless, by analyzing a very small library of only 324 different protein sequences we were able to recapitulate the wild-type affinity for the Hsp70 peptide in novel-binding site.
In this work we illustrate a general synergistic approach for designing novel binding affinities. We have successfully combined protein design and selection to redesign the binding site of the TPR1 protein. In the future, we anticipate the application of the methodology presented here for creation of new protein–protein interfaces for a variety of practical applications.
Funding
This work was supported in part by HFSF RGP0044/2007-C and NIH R01 GM080515-03 grants.
Acknowledgements
We thank Sarel Fleishman and David Baker for their interest and for advice and discussions about Rosetta modeling, and Alicia Morgan and James Lee for their contribution to computational and experimental work. We also thank Aitziber Lopez Cortajarena, Robielyn Ilagan, Lenka Kundrat and Robert Collins for valuable discussion and comments on the manuscript.
Footnotes
Edited by Valerie Daggett
References
- Binz H.K., Pluckthun A. Curr. Opin. Biotechnol. 2005;16:459–469. doi: 10.1016/j.copbio.2005.06.005. doi:10.1016/j.copbio.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Binz H.K., Amstutz P., Kohl A., Stumpp M.T., Briand C., Forrer P., Grutter M.G., Pluckthun A. Nat. Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. doi:10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- Binz H.K., Amstutz P., Pluckthun A. Nat. Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. doi:10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- Cortajarena A.L., Regan L. Protein Sci. 2006;15:1193–1198. doi: 10.1110/ps.062092506. doi:10.1110/ps.062092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortajarena A.L., Kajander T., Pan W., Cocco M.J., Regan L. PEDS. 2004;17:399–409. doi: 10.1093/protein/gzh047. [DOI] [PubMed] [Google Scholar]
- Cortajarena A.L., Wang J., Regan L. FEBS J. 2010;277:1058–66. doi: 10.1111/j.1742-4658.2009.07549.x. [DOI] [PubMed] [Google Scholar]
- Cortajarena A.L., Yi F., Regan L. ACS Chem. Biol. 2008;3:161–166. doi: 10.1021/cb700260z. doi:10.1021/cb700260z. [DOI] [PubMed] [Google Scholar]
- D'Andrea L., Regan L. Trends Biochem. Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. doi:10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Das R., Baker D. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. doi:10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Das A.K., Cohen P.W., Barford D. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. doi:10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove T.Z., Cortajarena A.L., Regan L. Curr. Opin. Struct. Biol. 2008;18:507–515. doi: 10.1016/j.sbi.2008.05.008. doi:10.1016/j.sbi.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackrel M.E., Cortajarena A.L., Liu T.Y., Regan L. ACS Chem Biol. 2009 doi: 10.1021/cb900272j. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Jackrel M.E., Valverde R., Regan L. Protein Sci. 2009;18:762–774. doi: 10.1002/pro.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., et al. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. doi:10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanicolas J., Kuhlman B. Curr. Opin. Struct. Biol. 2009;19:458–463. doi: 10.1016/j.sbi.2009.07.005. doi:10.1016/j.sbi.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajander T., Cortajarena A.L., Main E.R., Mochrie S.G., Regan L. J. Am. Chem. Soc. 2005;127:10188–10190. doi: 10.1021/ja0524494. doi:10.1021/ja0524494. [DOI] [PubMed] [Google Scholar]
- Kajander T., Sachs J.N., Goldman A., Regan L. J. Biol. Chem. 2009;284:25364–25374. doi: 10.1074/jbc.M109.033894. doi:10.1074/jbc.M109.033894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J.R., Tugendreich S., Hieter P. Trends Biochem. Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. doi:10.1016/S0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- Magliery T.J., Regan L. J. Mol. Biol. 2004;343:731–745. doi: 10.1016/j.jmb.2004.08.026. doi:10.1016/j.jmb.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Magliery T.J., Regan L. BMC Bioinformatics. 2005;6:240. doi: 10.1186/1471-2105-6-240. doi:10.1186/1471-2105-6-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliery T.J., Wilson C.G.M., Pan W.L., Mishler D., Ghosh I., Hamilton A.D., Regan L. J. Am. Chem. Soc. 2005;127:146–157. doi: 10.1021/ja046699g. doi:10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- Mandell D.J., Kortemme T. Nat. Chem. Biol. 2009;5:797–807. doi: 10.1038/nchembio.251. doi:10.1038/nchembio.251. [DOI] [PubMed] [Google Scholar]
- Rothlisberger D., et al. Nature. 2008;453:190–195. doi: 10.1038/nature06879. doi:10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F.U., Moarefi I. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. doi:10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Skerra A. FEBS J. 2008;275:2677–2683. doi: 10.1111/j.1742-4658.2008.06439.x. doi:10.1111/j.1742-4658.2008.06439.x. [DOI] [PubMed] [Google Scholar]
- Thyme S.B., Jarjour J., Takeuchi R., Havranek J.J., Ashworth J., Scharenberg A.M., Stoddard B.L., Baker D. Nature. 2009;461:1300–1304. doi: 10.1038/nature08508. doi:10.1038/nature08508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., Doudevski I., Regan L. Protein Sci. 2010;19:19–25. doi: 10.1002/pro.278. [DOI] [PMC free article] [PubMed] [Google Scholar]