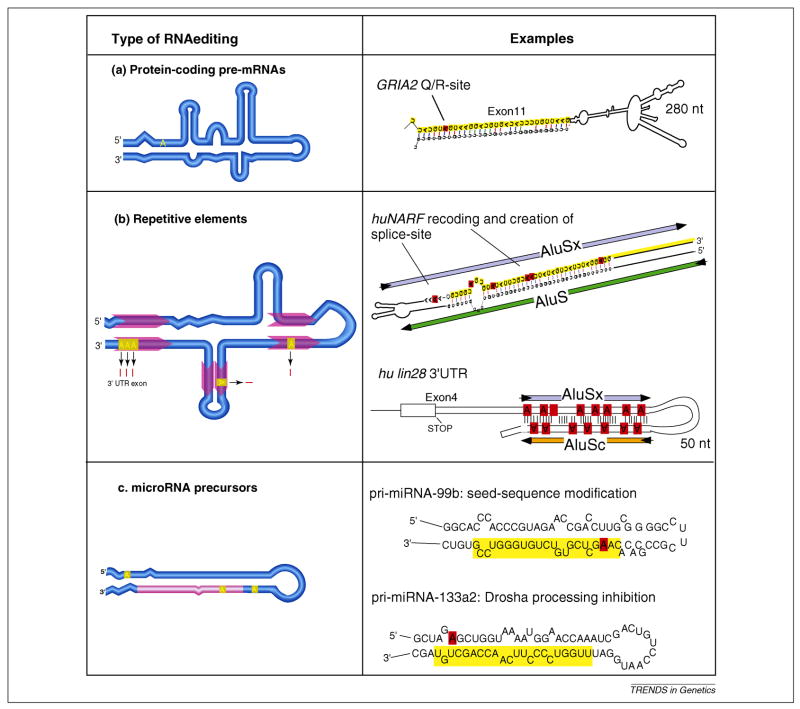
Figure 1.
The three major types of A-to-I RNA editing targets and their fates. Panels on the left show schematic of RNA secondary structures highlighting translated exon sequence (dark blue box), untranslated exon sequence (light blue boxes) location of repetitive sequence elements (red arrows), non-coding and intronic RNA sequence (light blue lines) and location of mature miRNA sequence (light red line). (a) Pre-mRNA editing of protein-coding genes with composite RNA secondary structure leads to highly site-selective recoding if it affects a non-synonymous codon site. For example, the glutamate receptor subunit GRIA2 exon 11 Q/R site 2 forms an experimentally validated secondary RNA structure between exon 11 (marked in yellow) and intron 11. (b) Pairs of repetitive elements, such as primate Alus located in coding or non-coding exons or introns can generate RNA secondary structures targeted by the RNA editing machinery. For example, editing of the intramolecular RNA fold between two Alu elements in human nuclear prelamin A recognition factor (NARF), causes recoding within the Alu-exon (marked in yellow) and leads to the creation of the 3′-splice consensus site upstream of the Alu-exon, thereby regulating alternative splicing of this exon. In the case of human lin28, extensive RNA editing within its non-coding, 3′-untranslated region mediated by a pair of Alu-elements, leads to the nuclear retention of the mRNA. (c) The characteristic secondary structure of pre-miRNAs is a frequent target of ADARs. For example, pri-miRNA-99b editing alters a nucleotide within the seed of the mature miRNA (marked in yellow and edited position highlighted in red) and therefore has the potential to alter the target interaction profile of this miRNA 26, whereas the modification of an adenosine outside of the mature miRNA region in pri-miRNA-133a2 causes a change in the processing rate by the RNAse Drosha 26.