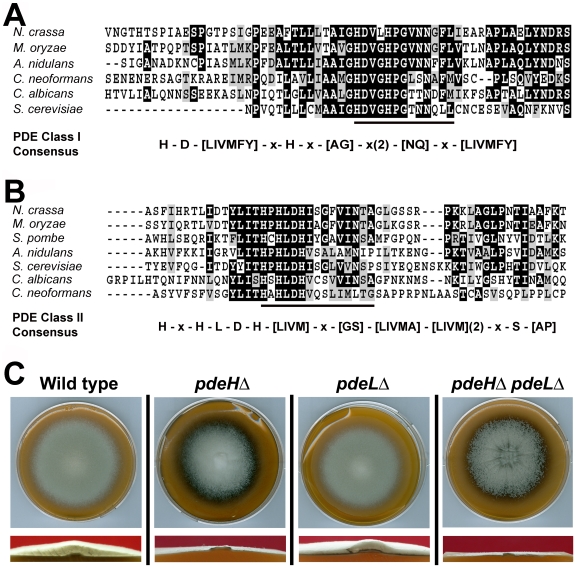
Figure 1. Identification and deletion analysis of cAMP phosphodiesterase genes in M. oryzae.
(A) Sequence alignment of the predicted active sites of the high-affinity cAMP phosphodiesterase from N. crassa (NCU00478), M. oryzae (MGG_05664), A. niger (AN2740.2), S. cerevisiae (CAA99689), C. albicans (AAM89252) and C. neoformans (AY874131). The conserved PDE Class I consensus is represented below. (B) Comparative alignment of the region containing the predicted signature sequence of the low-affinity cAMP phosphodiesterase from N. crassa (NCU00237), M. oryzae (MGG_07707), S. pombe (CAA20842), A. niger (AN0829.2), S. cerevisiae (CAA64139), C. albicans (XP720545) and C. neoformans (AY864841). The Class II PDE consensus sequence is underlined and depicted below in detail. S. pombe lacks the high affinity PDE variant. The multiple sequence alignment was performed using Clustal W and Boxshade (http://bioweb.pasteur.fr/seqanal/interfaces/boxshade.html). The conserved amino acid residues are shaded black, whereas similar residues are shown in gray. (C) PdeH is necessary for proper aerial hyphal growth. Morphology of the wild-type, pdeHΔ, pdeLΔ or the pdeHΔ pdeLΔ colonies. The indicated strains were grown in the dark on prune agar medium for a week and photographed (Upper panel). The lower panels show the cross sections of the above colonies at near-median planes. The pdeHΔ and the pdeHΔ pdeLΔ are dramatically reduced in aerial hyphal growth.