Figure 3.
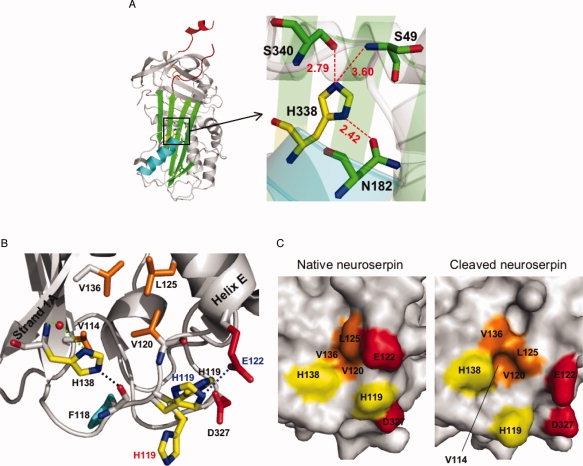
A: Schematic view of native neuroserpin (3FGQ).37 The reactive center loop is shown in red at the upper pole of the protein. The distances (in Å) between His338 and nearby residues in the shutter domain are shown (represented by the box on the left panel). B: Schematic view showing the interactions between strand 1A and helix E at the lower pole of native neuroserpin (3FGQ). The side chains of histidines (His138 and His119: yellow), the hydrophobic residues (Val114, Val120, Leu125, and Val136: orange), Asp327 (red), and Phe118 (cyan) are highlighted. His138 interacts with a water molecule (red sphere) by two hydrogen bonds (green dotted line), and the distance between NE2-His138 and O=C—Phe118 (black dotted line) is 2.50 Å. The distance between ND1-His119 and OD2-Asp327 is 2.64 Å. There is a π–π stacking interaction between the imidazole ring of His119 and O=C—Val120. For comparison, reactive loop-cleaved neuroserpin (3F02)42 and native neuroserpin obtained from different crystallizing conditions (3F5N)42 are superimposed, and His119 and Glu122 are shown with red (3F02) or blue letters (3F5N). The distance between NE2-His119 and OE1-Glu122 in 3F5N (blue dotted line) is 3.09 Å. C: Molecular surface of native neuroserpin (3FGQ: left) and reactive loop-cleaved neuroserpin (3F02: right). The side chains of His138, His119, Glu122, Asp327 and the hydrophobic residues (Val114, Val120, Leu125, and Val136) are highlighted with the same color as in panel B. Val114 is fully buried in native neuroserpin. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
