Figure 2.
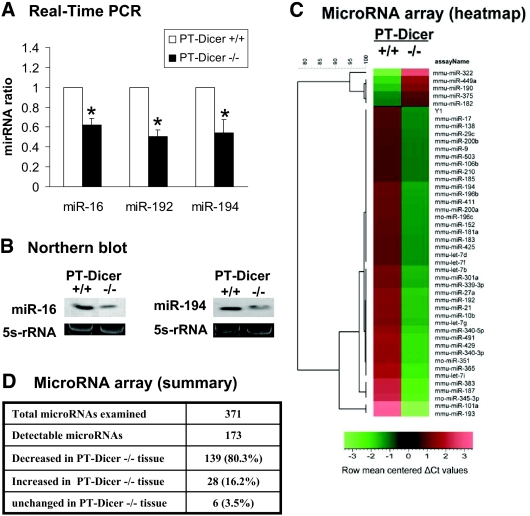
Depletion of miRNAs from renal cortical tissues in PT-Dicer−/− mice. Total RNA is isolated from renal cortical tissues of PT-Dicer+/+ and PT-Dicer−/− mice. (A) Real-time PCR analysis of miR-16, miR-192, and miR-194. Real-time PCR is performed using the Taqman miRNA assay kit as described in the Concise Methods section. The value of each miRNA is normalized by the signal of snoRNU202, an internal control. The normalized values of PT-Dicer+/+ samples are arbitrarily set as 1; data of PT-Dicer−/−samples are expressed as mean ± SD (n = 3). *Significantly different from the PT-Dicer+/+ values. (B) Northern blot analysis of miR-16 and miR-194. Total RNA (10 μg per lane) is analyzed by Northern blotting as described in the Concise Methods section using a p32-labeled probe of miR-16 or miR-194. 5s-rRNA is shown as an RNA loading control. (C) Representative heat map of microRNA microarray analysis. Total RNA samples isolated from renal cortical tissues of PT-Dicer+/+ and PT-Dicer−/− mice are subjected to microRNA microarray analysis. The ΔCt values of all miRNAs are used to generate the heat map. (D) Summary of microRNA microarray results. The microarray included 371 miRNA species, 173 of which were detectable in the renal cortical tissues. The value of a specific miRNA from the PT-Dicer−/− sample is compared with that of PT-Dicer+/+ sample to show whether the miRNA is decreased, increased, or unchanged. The results are representative of duplicate analyses.