Abstract
Electrophoretic surveys of hemoglobin (Hb) polymorphism in house mice from South Asia and the Middle East have revealed that two alternative β-globin haplotypes, Hbbd and Hbbp, are often present at intermediate frequencies in geographically disparate populations. Both haplotypes harbor two functionally distinct β-globin paralogs, HBB-T1 (which encodes the β-chain subunits of the major Hb isoform) and HBB-T2 (which encodes the β-chains of the minor Hb isoform). The Hbbd and Hbbp haplotypes share identical HBB-T1 alleles, but products of the alternative HBB-T2 alleles (dminor and pminor) are distinguished by two amino acid substitutions. To investigate the possible adaptive significance of the Hbbd/Hbbp polymorphism we conducted a population genetic analysis of the duplicated β-globin genes of Indian house mice (Mus castaneus) in conjunction with experimental studies of Hb function in inbred strains of mice that carry the alternative Hbbd and Hbbp haplotypes. The main objectives of this study were (i) to characterize patterns of nucleotide polymorphism and linkage disequilibrium in the duplicated β-globin genes of M. castaneus, (ii) to test the hypothesis that the Hbbd and Hbbp haplotypes are maintained as a balanced polymorphism, and (iii) to assess whether allelic differences in the alternative minor Hb isoforms (dminor and pminor) are associated with different O2-binding properties. A multilocus analysis of polymorphism and divergence revealed that levels of diversity at the HBB-T2 gene exceeded neutral expectations, and reconstructed haplotype networks for both β-globin paralogs revealed extensive allele sharing with several other closely related species of Mus. However, despite this suggestive evidence for balancing selection, O2-equilibrium curves revealed no discernible functional differences between red cell lysates containing the dminor and pminor Hb isoforms. If the dminor and pminor alleles are maintained as a balanced polymorphism, our results indicate that the associated fitness variance is not directly related to respiratory functions of Hb.
BALANCING selection at a particular locus is expected to produce elevated levels of nucleotide diversity and linkage disequilibrium (LD) due to the partitioning of sequence variation between unusually long-lived allele classes (Hudson and Kaplan 1988; Charlesworth et al. 2003; Charlesworth 2006). Although elevated levels of nucleotide polymorphism at a particular gene may provide suggestive evidence for a history of balancing selection, conclusive inferences regarding the selective maintenance of allelic polymorphism ultimately require experimental evidence that the alternative alleles are functionally distinct. The well-characterized β-globin polymorphism in house mice (genus Mus) represents a system where it is possible to integrate evolutionary and functional approaches to evaluate the role of balancing selection in maintaining protein polymorphism.
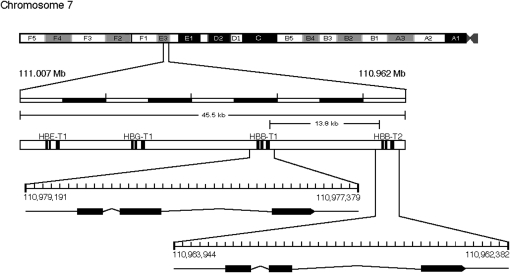
The two tandemly duplicated β-globin genes of house mice, HBB-T1 and HBB-T2, encode the β-chain subunits of adult hemoglobin (Hb) and are separated by ∼12–15 kb on chromosome 7 (Hoffmann et al. 2008; Figure 1). Five main classes of β-globin haplotypes have been characterized in natural populations of house mice: Hbbd, Hbbp, Hbbs, Hbbw1, and Hbbw2 (Sato et al. 2008; Runck et al. 2009). Electrophoretic surveys of β-globin polymorphism in natural populations of Mus musculus and M. domesticus have revealed that the Hbbd and Hbbs haplotypes are consistently present at intermediate frequencies in population samples from across the species' range, a pattern that is not paralleled at other unlinked autosomal genes (Petras 1967; Selander et al. 1969a,b; Selander and Yang 1969; Berry and Murphy 1970; Wheeler and Selander 1972; Myers 1974; Berry and Jakobson 1975; Berry and Peters 1975, 1977, 1981; Berry et al. 1978; Sage 1981; Petras and Topping 1983; Sage et al. 1986). This striking uniformity of two-locus haplotype frequencies has led a number of authors to conclude that the polymorphism may be maintained by some form of balancing selection (reviewed by Berry 1978). More recently, surveys of nucleotide variation at the two β-globin paralogs have revealed patterns consistent with the idea that the Hbbd and Hbbs haplotypes are maintained as a balanced polymorphism. First, high levels of silent site polymorphism at the HBB-T1 and HBB-T2 genes of M. domesticus exceeded neutral expectations on the basis of levels of interspecific divergence (Storz et al. 2007). Second, the Hbbd and Hbbs haplotypes are shared among multiple Eurasian species in the subgenus Mus, indicating that the time to the most recent common ancestor of the two haplotypes may predate multiple speciation events (Runck et al. 2009). However, transpecific polymorphism appears to be relatively common in house mice due to incomplete lineage sorting and introgressive hybridization (e.g., Salcedo et al. 2007; Geraldes et al. 2008). Thus, central assumptions of standard neutrality tests may often be violated, making it especially important to use direct, experimental approaches to test hypotheses about balancing selection.
Figure 1.—
Genomic structure of the β-globin gene family of the house mouse (BALB/c strain) on chromosome 7.
The Hbbd haplotype harbors two distinct β-globin paralogs that are distinguished from one another by 9 amino acid substitutions. The more highly expressed HBB-T1 gene encodes the β-chain subunits of the major Hb isoform (isoHb), dmajor, whereas HBB-T2 encodes the β-chains of the minor isoHb, dminor (Hutton et al. 1962; Gilman 1972, 1974; Whitney 1977). In contrast to the two divergent β-globin paralogs on the Hbbd haplotype, the Hbbs haplotype harbors two β-globin paralogs that are identical in sequence due to a history of HBB-T1→HBB-T2 gene conversion (Erhart et al. 1985; Storz et al. 2007). Consequently, mice that are homozygous for the Hbbs haplotype synthesize a single β-chain isoHb during postnatal life. The Hbbd and Hbbs haplotypes are distinguished by 3 amino acid substitutions at HBB-T1 and 11 amino acid substitutions at HBB-T2 (Erhart et al. 1985; Storz et al. 2007). One of the most salient differences between Hbbd and Hbbs is that β-chain products of HBB-T1 and HBB-T2 on the Hbbd haplotype (dmajor and dminor, respectively) contain a derived cysteine residue at site 13, which appears to play an important role in intraerythrocyte glutathione metabolism and nitric oxide metabolism (Miranda 2000; Giustarini et al. 2006; Hempe et al. 2007).
A polymorphism involving the Hbbd and Hbbp haplotypes has been documented in house mice from South Asia and the Middle East (Minezawa et al. 1979; Ritte and Neufeld 1982; Miyashita et al. 1985; Kawashima et al. 1995), which exhibits some similarities to the Hbbd/Hbbs polymorphism in house mice from Europe and the Americas. Similar to the pattern described for the Hbbd/Hbbs polymorphism, the Hbbd and Hbbp haplotypes are present at intermediate frequencies in geographically disparate population samples from across the range of the Asian house mouse M. castaneus (Miyashita et al. 1985; Kawashima et al. 1995). As in the case with the Hbbd haplotype, the HBB-T1 and HBB-T2 paralogs on the Hbbp haplotype encode the β-chain subunits of the major and minor isoHbs (pmajor and pminor), respectively. The Hbbd and Hbbp haplotypes share identical HBB-T1 sequences (dmajor = pmajor), but they are distinguished by two amino acid substitutions at HBB-T2: 22Glu→Ala and 23Val→Ile (dminor→pminor, in both cases). The sequence identity between HBB-T1 alleles on the Hbbd and Hbbp haplotypes reflects the fact that Hbbp is a recombinant chromosome that was produced by intergenic crossing over between Hbbd and Hbbw1 parental chromosomes (Ueda et al. 1999; Sato et al. 2008; Runck et al. 2009). Consequently, Hbbp carries an HBB-T1 allele derived from Hbbd and an HBB-T2 allele derived from Hbbw1. In contrast to the structural differences between products of Hbbd and Hbbs in M. domesticus and M. musculus, the β-chain isoHbs produced by Hbbd and Hbbp do not differ in cysteine content. If there is some functionally significant difference between the β-chain isoHbs produced by the Hbbd and Hbbp haplotypes it must be related to the two amino acid differences that distinguish the minor isoHbs, dminor and pminor.
To investigate the possible adaptive significance of the Hbbd/Hbbp polymorphism we conducted a population genetic analysis of the HBB-T1 and HBB-T2 genes in a natural population of M. castaneus from northern India. In conjunction with this evolutionary analysis of sequence variation, we also conducted an experimental study of Hb function in inbred strains of mice that carry each of the alternative two-locus β-globin haplotypes. The main objectives of this study were (i) to characterize patterns of nucleotide polymorphism and LD in the duplicated β-globin genes of M. castaneus, (ii) to test the hypothesis that the Hbbd and Hbbp haplotypes are maintained as a balanced polymorphism, and (iii) to assess whether allelic differences in the alternative minor isoHbs (dminor and pminor) are associated with different O2-binding properties.
MATERIALS AND METHODS
Samples for the population genetic analysis:
Our survey of nucleotide variation at the HBB-T1 and HBB-T2 genes was based on a total of 24 M. castaneus specimens that were collected by B. Harr in northern India [for details, see Baines and Harr (2007) and Geraldes et al. (2008)]. The mice used in our survey were collected from four closely situated localities within a 130 km radius that spanned the border between the Indian states of Himachal Pradesh and Uttarakhand. Mice from the Indian subcontinent have been referred to as M. bactrianus by some authors (see Boursot et al. 1993). Following Baines and Harr (2007) and Geraldes et al. (2008), we refer to the specimens from northern India as M. castaneus.
PCR, cloning, and sequencing:
Genomic DNA was extracted from ethanol-preserved liver tissue of each mouse using DNeasy kits (Qiagen, Valencia, CA). We cloned and sequenced both alleles of the HBB-T1 and HBB-T2 genes in the full sample of mice (48 experimentally phased sequences per gene, 96 sequences total). We therefore obtained complete diploid genotypes for both HBB paralogs in each mouse, and we were able to determine the exact haplotype phase of all heterozygous sites. Amplification of the complete coding regions of HBB-T1 and HBB-T2 followed established protocols (Runck et al. 2009). For HBB-T1, the sequenced fragment was 1809 bp in length, including 329 bp of 5′ flanking sequence and 276 bp of 3′ flanking sequence. For HBB-T2, the sequenced fragment was 1793 bp, including 338 bp of 5′ flanking sequence and 300 bp of 3′ flanking sequence. PCR products were cloned into pCR4-TOPO vector following the manufacturer's protocols (Invitrogen, Carlsbad, CA). For each species, we sequenced a total of 8–10 colonies per gene using the vector primers T3 and T7 (54° annealing). Automated DNA sequencing was performed on an ABI 3730 capillary sequencer using Big Dye chemistry (Applied Biosystems, Foster City, CA). Sequences were deposited in GenBank under the accession numbers GU057161–GU057256.
Analysis of DNA sequence variation:
HBB-T1 and HBB-T2 sequences were aligned and assembled into contigs using ClustalX (Thompson et al. 1997) and Sequencher (Gene Codes, Ann Arbor, MI). For the purpose of reconstructing the evolutionary history of the HBB-T1 and HBB-T2 sequences from Indian M. castaneus, and to test for evidence of transpecific polymorphism, we reconstructed haplotype networks that included previously published sequence data from the orthologous β-globin genes of four additional species of Mus (M. domesticus, M. macedonicus, M. musculus, and M. spicilegus) and an additional M. castaneus specimen from Thailand (Storz et al. 2007; Runck et al. 2009). M. castaneus, M. domesticus, M. macedonicus, M. musculus, and M. spicilegus are each known to carry the Hbbd haplotype, and M. castaneus and M. musculus are also known to carry the Hbbp haplotype (Runck et al. 2009). Haplotype networks were constructed using equally weighted characters in the program NETWORK v4.5.1 (www.fluxus-engineering.com). To ensure that a full median network was calculated, we set the weighted genetic distance measure (ɛ) to equal the maximum number of pairwise differences between haplotypes at each paralog (ɛ = 10 for HBB-T1 and ɛ = 11 for HBB-T2). We used the maximum parsimony option to remove unnecessary median vectors in the full median network (Polzin and Daneschmand 2003).
After binning HBB-T1 and HBB-T2 amino acid sequences into discrete allele classes, we calculated Weir and Cockerham's (1984) estimator of the inbreeding coefficient, F. To assess whether observed genotype frequencies deviated from Hardy–Weinberg equilibrium we used the exact test of Guo and Thompson (1992). We used a similar Markov-chain contingency table method to test the null hypothesis that genotypes at one β-globin gene were independent of genotypes at the other gene.
Summary statistics of nucleotide polymorphism and LD were computed with the programs SITES (Hey and Wakeley 1997) and DnaSP v5 (Librado and Rozas 2009). To detect intragenic recombination within each of the two β-globin paralogs, we used the four-gamete test of Hudson and Kaplan (1985) to estimate RM, the minimum number of recombination events in the history of the sample. We computed two different measures of DNA sequence variation: nucleotide diversity, π (the average number of pairwise differences between sequences) and Watterson's (1975) estimator of the scaled mutation rate, θW (4Nu, where N is the effective population size and u is the mutation rate per nucleotide). We used variation at coding and noncoding sites to compute Hey and Wakeley's (1997) estimator of the population recombination rate, γ. We used the method of Betrán et al. (1997) to test for evidence of gene conversion between the two β-globin paralogs.
As a measure of LD between pairs of nucleotide polymorphisms, we used the squared allele-frequency correlation, r2, and we used Fisher's exact test to determine the probability of obtaining estimates of LD that were more extreme than the observed values under the null hypothesis of linkage equilibrium. The analysis was based on biallelic nucleotide polymorphisms where the minor allele was present at least twice in the sample. We used nonlinear regression to model the decay of intragenic LD as a function of physical distance under a model of recombination–drift equilibrium that incorporated mutation (Hill and Weir 1988). Specifically, we used a nonlinear regression model based on the Gauss–Newton algorithm, as implemented in the nls function of the R statistical computing package (www.r-project.org). To summarize the effects of recombination on intragenic LD, we also computed the ZZ test statistic of Rozas et al. (2001), which measures the difference between the average r2 between adjacent nucleotide polymorphisms and the average of pairwise r2 values across the entire gene. To compute confidence intervals of the ZZ test statistic we conducted 10,000 coalescent simulations (with no recombination) that were conditioned on the observed number of segregating sites.
Using data on silent site polymorphism at both β-globin paralogs, we conducted neutrality tests on the basis of two different summary statistics: Tajima's (1989) D, which provides a measure of the site-frequency distribution, and Kelly's (1997) ZnS, which provides a measure of intralocus LD. We obtained critical values for each test statistic by conducting coalescent simulations as described above.
To assess whether relative levels of polymorphism and divergence deviated from neutral expectations, we conducted a multilocus Hudson–Kreitman–Aguadé (HKA) test (Hudson et al. 1987). The analysis included polymorphism data from the HBB-T1 and HBB-T2 genes in addition to previously published data from 8 loci that were sequenced in the same panel of Indian M. castaneus specimens (Geraldes et al. 2008). The set of effectively unlinked reference loci included four autosomal genes (Chrng, Med19, Prpf3, and Clcn6), two X-linked genes (G6pdx and Ocr1), one Y-linked gene (Jarid 1d), and the mtDNA control region. For each of the 10 loci we used orthologous sequences from M. caroli (Geraldes et al. 2008; Runck et al. 2009) to estimate locus-specific values of Dxy, the average pairwise sequence divergence between species (Nei and Kumar 2000). Since the 5′ end of the M. caroli HBB-T1 gene has experienced a history of gene conversion by HBB-T2 (Runck et al. 2009), we treated the 624-bp conversion tract as missing data. Thus, for the HBB-T1 gene, our estimate of net divergence between M. castaneus and M. caroli was based on 1369 bp of orthologous sequence that spanned intron 2, exon 3, and the 3′-UTR. For the mtDNA control region, the estimate of net sequence divergence between M. castaneus and M. caroli was corrected for multiple hits (see Geraldes et al. 2008). We jointly estimated neutral parameters from all 10 loci to obtain expected values for the HKA test, and we then used the resultant estimates to conduct coalescent simulations using the HKA program (http://genfaculty.rutgers.edu/hey/software).
Experimental analysis of Hb function:
To test for functional differences between the β-chain isoHbs produced by the Hbbd haplotype (dmajor and dminor) and the Hbbp haplotype (pmajor and pminor), we measured O2-binding properties of hemolysates from the BALB/c and MSM/s inbred strains. Blood samples from BALB/c (which is homozygous for Hbbd) and MSM/s (which is homozygous for Hbbp) were obtained from the Jackson Lab (Bar Harbor, ME). Blood samples from the MSM/s strain were procured under a material transfer agreement with the National Institute of Genetics (Mishima, Japan).
Hemolysates were prepared according to standard methods and were stripped of organic phosphates and other ionic cofactors as described previously (Storz et al. 2009). The isoHb composition of hemolysates from each mouse strain was confirmed by using thin-layer isoelectric focusing (PhastSystem, GE Healthcare Biosciences, Piscataway, NJ). Using a modified diffusion chamber, O2 equilibria of Hb solutions were measured in 10 mm HEPES buffer, pH 7.4, at constant temperature, 37°. The met-Hb enzymatic reducing system of Hayashi et al. (1973) was used to prevent oxidation of ferrous heme. Changes in the absorbance of Hb solutions were recorded in conjunction with stepwise changes in the partial pressure of O2 [PO2] inside the chamber (prepared using cascaded Wösthoff gas-mixing pumps; Weber 1981, 1992; Weber et al. 2004). Values of P50 (the PO2 at 50% oxygenation of the heme groups) and n50 (Hill's cooperativity coefficient at P50) were interpolated from linear Hill plots (log ([OxyHb]/[Hb]) vs. log PO2). The P50 values for the stripped (i.e., cofactor-free) hemolysates provide an inverse measure of the intrinsic Hb–O2 binding affinities of the major and minor isoHbs occurring in their natural relative concentrations. To test for differences in cofactor sensitivity between the d- and p-type Hbs, we measured O2-equilibrium curves for each sample in the absence of allosteric cofactors (stripped hemolysates), in the presence of 2,3-diphosphoglycerate (DPG), in the presence of Cl− ions (added as potassium chloride, KCl), and in the presence of both cofactors [(Cl−), 0.10 m; (NaHEPES), 0.1 m; DPG/Hb tetramer ratio, 2.0; (Heme), 0.16 mm].
RESULTS
Genetic variation:
Our survey of nucleotide polymorphism in the HBB-T1 and HBB-T2 genes revealed that Indian house mice segregate two main β-globin haplotypes that appear to be referable to Hbbd and Hbbp. The survey of nucleotide polymorphism at HBB-T1 revealed the presence of three distinct allele classes that were present at roughly equal frequencies (Table 1). Allele classes T1A, T1B, and T1C were defined by the following two-site amino acid combinations at residue positions 16 and 20: T1A = Gly-Ser, T1B = Gly-Ala, and T1C = Ala-Ser. The amino acid sequence of the T1A allele is identical to the canonical dmajor and pmajor β-globin sequences that were originally characterized in the BALB/c and AU/SsJ inbred strains, respectively (Gilman 1972, 1974; Erhart et al. 1985). The T1B and T1C alleles differ from the canonical dmajor/pmajor sequence by single amino acid substitutions. The survey of nucleotide variation at HBB-T2 revealed the presence of two distinct allele classes, referable to dminor and pminor, that were also present at roughly equal frequencies (Table 1). The HBB-T2 allele classes were distinguished by the following two-site amino acid combinations at residue positions 22 and 23: dminor = Glu-Ile and pminor = Ala-Val. Relative to the expected genotype frequencies at Hardy–Weinberg equilibrium, both β-globin genes exhibited a highly significant deficit of heterozygotes (HBB-T1: F = 0.573, P < 0.0001; HBB-T2: F = 0.676, P = 0.0011). A more detailed inspection of genotype frequencies revealed that the observed heterozygote deficits were mainly attributable to population structure. Although the dminor and pminor alleles were present at respective frequencies of 0.54 and 0.46 in the pooled sample, the three more northern samples from Himachal Pradesh were enriched for pminor (frequency = 0.86, n = 11 mice) and the more southern sample from Uttarakhand was enriched for dminor (frequency = 0.88, n = 13 mice).
TABLE 1.
Amino acid variation among HBB-T1 and HBB-T2 alleles in Indian house mice
| Gene | Allele class | Frequency | Amino acid | |||
|---|---|---|---|---|---|---|
| 16 | 20 | 22 | 23 | |||
| HBB-T1 | T1A | 0.27 | Gly | Ser | Glu | Val |
| T1B | 0.35 | Gly | Ala | Glu | Val | |
| T1C | 0.38 | Ala | Ser | Glu | Val | |
| HBB-T2 | dminor | 0.54 | Gly | Ser | Glu | Ile |
| pminor | 0.46 | Gly | Ser | Ala | Val | |
Both HBB-T1 and HBB-T2 exhibited high levels of nucleotide diversity at silent sites (π = 0.0051 and 0.0110, respectively; Table 2). The estimates of θW for the HBB-T1 and HBB-T2 genes (θW = 0.0071 and 0.0148, respectively; Table 2) exceeded the upper range of values for the seven unlinked nuclear loci that were sequenced in the same panel of mice (θW = 0.0014–0.0062, where values for the X- and Y-linked loci were multiplied by 4/3 and 4, respectively, to account for differences in effective population size; Geraldes et al. 2008). The multilocus HKA test was highly significant due to a higher-than-expected level of polymorphism at the HBB-T2 gene (χd.f.=92 = 34.661, P = 0.00007; Table 3). The test also remained highly significant when the analysis was restricted to the six autosomal loci (χd.f.=52 = 22.644, P = 0.00030). The HBB-T1 and HBB-T2 genes both exhibited an excess of low-frequency polymorphisms, as indicated by negative values for Tajima's D, but the skews in the site-frequency distributions were not statistically significant in either case (Table 2).
TABLE 2.
Summary of nucleotide polymorphism and linkage disequilibrium at the HBB-T1 and HBB-T2 genes of Indian house mice
| Gene | Allele class | Length (bp) | N | S | H | HD | π(silent) | θw(silent) | Tajima's D | k | Rm | γ/bp | ZnS | ZZ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HBB-T1 | All | 1815 | 48 | 61 | 24 | 0.946 | 0.00505 | 0.00708 | −0.887 | 10.17 | 5 | 0.00975 | 0.0936 | 0.1587* |
| T1A | 1816 | 13 | 9 | 5 | 0.628 | 0.00112 | 0.00160 | −1.188 | 2.03 | |||||
| T1B | 1816 | 17 | 10 | 7 | 0.816 | 0.00156 | 0.00163 | −0.148 | 2.83 | |||||
| T1C | 1814 | 18 | 44 | 12 | 0.954 | 0.00485 | 0.00673 | −1.141 | 8.98 | |||||
| HBB-T2 | All | 1802 | 48 | 125 | 35 | 0.986 | 0.01101 | 0.01475 | −0.8377 | 21.38 | 8 | 0.01052 | 0.0739 | 0.1093* |
| dminor | 1804 | 26 | 78 | 18 | 0.966 | 0.00692 | 0.01060 | −1.348 | 12.53 | |||||
| pminor | 1803 | 22 | 50 | 17 | 0.978 | 0.00440 | 0.00761 | −1.664 | 7.76 |
*P < 0.05. N, sampled chromosomes; S, total number of segregating sites; H, number of haplotypes; HD, haplotype diversity; and k, average number of nucleotide differences between sequences.
TABLE 3.
Multilocus HKA test involving the HBB-T1 and HBB-T2 genes and eight reference loci
| Polymorphism |
Divergence |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Locus | Chromosome | I | θ (2.5–97.5%) | S/ES | Var | Dev | D/ED | Var | Dev |
| HBB-T1 | 7 | 1.00 | 0.0056 (0.0038–0.0084) | 57/45.46 | 215.82 | 0.618 | 44/55.54 | 126.82 | 1.051 |
| HBB-T2 | 7 | 1.00 | 0.0082 (0.0056–0.0119) | 118/65.39 | 417.90 | 6.624 | 44/96.61 | 312.25 | 8.865 |
| Chrng | 1 | 1.00 | 0.0059 (0.0040–0.0093) | 61/55.44 | 308.90 | 0.100 | 42/47.56 | 99.80 | 0.309 |
| Med19 | 2 | 1.00 | 0.0057 (0.0039–0.0082) | 15/44.05 | 193.69 | 4.358 | 92/62.95 | 154.48 | 5.464 |
| Prpf3 | 3 | 1.00 | 0.0032 (0.0021–0.0046) | 31/35.25 | 132.57 | 0.136 | 55/50.75 | 110.25 | 0.164 |
| Clcn6 | 4 | 1.00 | 0.0058 (0.0039–0.0082) | 54/52.68 | 270.00 | 0.006 | 75/76.32 | 210.89 | 0.008 |
| G6pdx | X | 0.75 | 0.0037 (0.0025–0.0053) | 19/26.35 | 95.43 | 0.567 | 63/55.65 | 99.13 | 0.545 |
| Orc1 | X | 0.75 | 0.0056 (0.0038–0.0081) | 32/31.89 | 141.81 | 0.000 | 68/68.11 | 133.26 | 0.000 |
| Jarid 1d | Y | 0.25 | 0.0072 (0.0049–0.0105) | 4/13.56 | 40.99 | 2.232 | 68/58.44 | 64.72 | 1.413 |
| Control region | mtDNA | 0.25 | 0.0598 (0.0438–0.0803) | 33/53.93 | 338.90 | 1.292 | 329/308.07 | 482.64 | 0.907 |
χd.f.=92 = 34.661, P = 0.00007. I, inheritance scalar (1.00 for autosomal loci, 0.75 for X-linked loci and 0.25 for the Y-linked locus and mtDNA); S, observed number of segregating sites; ES, expected number of segregating sites; Var, variance under the model; Dev, deviation of each observed value from the model-based expectation; D, observed divergence to M. caroli; and ED, expected divergence.
Levels and patterns of LD:
In addition to the departure from Hardy–Weinberg genotype frequencies, we also observed a highly nonrandom association between genotypes at each of the two β-globin genes (χd.f.=22 = 19.807, P < 0.0001). This high level of intergenic LD is partly attributable to population structure, as the difference in allele frequencies between the northern and southern collection localities augments the covariance in allele frequencies between the two β-globin genes. At the HBB-T1 gene, Fisher's exact test revealed significant LD between 189 of 703 pairwise comparisons (56 of which remained statistically significant after Bonferroni correction), and at the HBB-T2 gene, significant LD was observed between 563 of 2485 such comparisons (304 of which remained significant after Bonferroni correction). Neither of the two β-globin genes exhibited an excess of intragenic LD, as indicated by nonsignificant ZnS values (Table 2). HBB-T1 and HBB-T2 both showed evidence for a history of intragenic recombination (Rm = 5 and 8, respectively), and we detected no evidence for interparalog gene conversion.
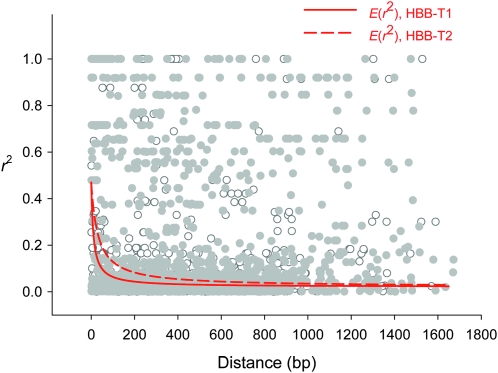
To assess whether the elevated polymorphism at HBB-T1 and HBB-T2 could plausibly be explained by the effects of balancing selection at a linked locus, we measured rates of decay of intralocus LD at each of the two β-globin genes. The rationale is that if LD decays to near background levels within each individual gene, then the observed allelic dimorphism at the HBB-T1 and HBB-T2 genes cannot be ascribed to the effects of selection at a linked locus. At both β-globin genes, mean r2 declined to <0.1 within 200 bp (Figure 2). Although a relatively small number of nonrandom associations persisted for site pairs separated by up to 1.5 kb, it is clear that LD does not extend very far into flanking chromosomal regions. Consistent with these results, estimated values of the ZZ test statistic were significantly positive for both HBB-T1 and HBB-T2 (Table 2), indicating that intragenic recombination has played an important role in randomizing pairwise associations between polymorphisms within each gene.
Figure 2.—
Rates of decay of gametic linkage disequilibrium within the HBB-T1 and HBB-T2 genes. The red lines show nonlinear regressions of r2 against physical distance using a mutation–recombination–drift model (see text for details). Open symbols denote r2 estimates for pairs of polymorphic sites in the HBB-T1 gene, and solid symbols denote estimates for pairs of polymorphic sites in the HBB-T2 gene.
Evolutionary relationships among house mouse β-globin sequences:
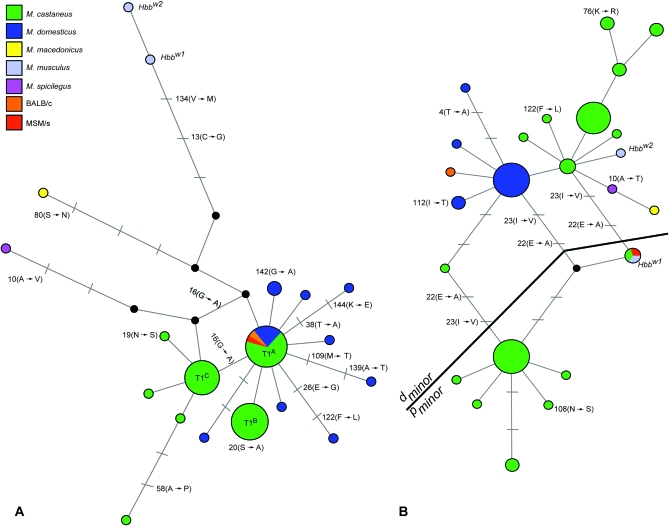
The minimum spanning network of HBB-T1 coding sequences revealed that many of the HBB-T1 alleles from M. castaneus were more similar to alleles from M. domesticus than they were to other HBB-T1 alleles from M. castaneus (Figure 3A). The Hbbw1 and Hbbw2 alleles of M. musculus (Kawashima et al. 1991; Sato et al. 2008) were the most distantly related sequences in the network of HBB-T1 alleles. Relative to HBB-T1, we observed an even greater degree of transpecific polymorphism at the HBB-T2 gene. The dminor alleles from M. castaneus were far more closely related to dminor alleles from M. domesticus, M. macedonicus, M. musculus, and M. spicilegus than they were to pminor alleles from M. castaneus (Figure 3B). Even within the dminor allele class, many dminor sequences from M. castaneus were more similar to those of M. domesticus and M. musculus than they were to other conspecific dminor sequences.
Figure 3.—
Median joining network showing relationships among β-globin coding sequences from M. castaneus and four other species of Mus that are known to segregate the Hbbd and Hbbp haplotypes (M. domesticus, M. macedonicus, M. musculus, and M. spicilegus). Haplotype networks for the HBB-T1 and HBB-T2 paralogs are shown in A and B, respectively. The size of each circle is proportional to the corresponding haplotype frequency. Inferred intermediate haplotypes are shown as black circles on branches connecting observed haplotypes. Branches between haplotypes indicate one mutational step, and tick marks denote additional steps. Amino acid changes in HBB-T1 sequences are shown in relation to the canonical dmajor/pmajor sequence from the BALB/c and MSM/s inbred strains, and amino acid changes in HBB-T2 are shown in relation to the canonical dminor sequence from BALB/c. The three allele classes at HBB-T1 are indicated in A. The black line in B separates haplotypes that are referable to the dminor and pminor allele classes.
Functional properties of house mouse Hbs:
To test for functional differences between the minor β-chain isoHbs produced by the Hbbd haplotype (dminor) and the Hbbp haplotype (pminor), we compared O2-binding properties of hemolysates from the BALB/c and MSM/s inbred strains. Hb–O2 affinity is modulated by various allosteric cofactors, particularly protons (responsible for the Bohr effect) and organic and inorganic anions, such as DPG and Cl− ions (Weber and Fago 2004; Storz and Moriyama 2008). In the red blood cells of most mammalian species, the binding of these effectors stabilize the low-affinity, deoxygenated “tense state” conformation of the Hb tetramer relative to the oxygenated “relaxed state,” thereby decreasing Hb–O2 affinity. Thus, even if the dminor and pminor Hbs exhibit similar intrinsic O2-binding affinities, differences in their cofactor sensitivities could still account for significant differences in blood–O2 affinity.
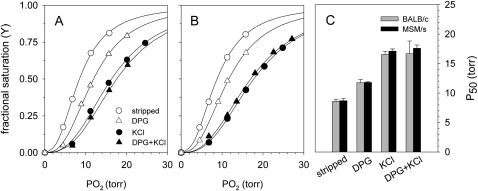
The comparison between hemolysates containing the dminor and pminor isoHbs revealed no significant difference in O2-binding properties (Figure 4). Across all treatments, the O2-affinities and cooperativity coefficients were nearly identical (Table 4). In summary, the O2-binding measurements revealed no discernible differences in the respiratory functions of dminor and pminor isoHbs under physiological conditions.
Figure 4.—
O2-equilibrium curves of stripped house mouse Hbs at pH 7.40 and 37° in the presence and absence of allosteric cofactors [(Cl−), 0.10 m; (NaHEPES), 0.1 m; DPG/Hb tetramer ratio, 2.0; (Heme), 0.16 mm] and in the presence of the met-Hb reductase system (Hayashi et al. 1973). Representative curves for the Hbs from two inbred strains, BALB/c (which is homozygous for Hbbd) and MSM/s (which is homozygous for Hbbp), are shown in A and B, respectively. C shows mean values (± SEM) of P50 (n = 3) measured under the above conditions and indicates that the β-chain isoHbs produced by the Hbbd haplotype and the Hbbp haplotype do not differ in intrinsic O2 affinity (as revealed by the comparison of stripped Hbs) or in sensitivity of Hb–O2 to the presence of allosteric cofactors such as DPG or Cl− ions.
TABLE 4.
O2 binding properties (mean ± SEM, n = 3) of stripped hemolysates from the BALB/c and MSM/s inbred strains (which are homozygous for the Hbbd and Hbbp β-globin haplotypes, respectively)
| Strain (β-haplotype) | Conditions | P50 (torr) | n50 |
|---|---|---|---|
| BALB/c (Hbbd) | Stripped | 8.44 ± 0.09 | 2.46 ± 0.05 |
| DPG | 11.88 ± 0.17 | 2.65 ± 0.09 | |
| KCl | 16.27 ± 0.30 | 2.68 ± 0.12 | |
| DPG + KCl | 17.53 ± 0.27 | 2.78 ± 0.13 | |
| MSM/s (Hbbp) | Stripped | 8.76 ± 0.04 | 2.40 ± 0.02 |
| DPG | 11.64 ± 0.07 | 2.50 ± 0.04 | |
| KCl | 17.74 ± 0.06 | 2.67 ± 0.02 | |
| DPG + KCl | 17.03 ± 0.27 | 2.56 ± 0.09 |
P50 and n50 values indicate the O2 tensions and cooperativity coefficients at half-saturation, respectively. O2 equilibria were measured in 0.1 m HEPES buffer at pH 7.40, 37°, in the absence of allosteric cofactors (stripped hemolysates), in the presence of DPG alone, in the presence of KCl alone, and in the presence of both cofactors [(Cl−), 0.10 m; DPG/Hb tetramer ratio, 2.0; (Heme), 0.16 mm].
DISCUSSION
Consistent with the results of electrophoretic surveys of house mice from other localities in South Asia (Miyashita et al. 1985; Kawashima et al. 1995), our survey of nucleotide polymorphism in the HBB-T1 and HBB-T2 genes revealed that Indian M. castaneus segregate two main β-globin haplotypes that are referable to Hbbd and Hbbp. Remarkably, many of the HBB-T1 and HBB-T2 alleles that we recovered in our sample of Indian M. castaneus were shared by other species of Mus. The transpecific polymorphism was especially pervasive at the HBB-T2 gene, as identical dminor and pminor alleles were shared among representatives of M. castaneus, M. domesticus, M. macedonicus, and M. spicilegus. A lesser degree of shared polymorphism among M. castaneus and M. domesticus was also documented at several of the other unlinked reference loci (Geraldes et al. 2008), which appears to reflect the combined effects of unsorted ancestral polymorphism and introgressive hybridization. Hybridization between M. castaneus and M. musculus has been documented in northern China and in Japan (Bonhomme et al. 1989; Boursot et al. 1993), and hybridization between M. castaneus and M. domesticus has been documented in California (Orth et al. 1998). At least in the case of M. castaneus, M. domesticus, and M. musculus, which are thought to have diverged from one another 350,000–900,000 years ago (She et al. 1990; Boursot et al. 1996; Suzuki et al. 2004; Salcedo et al. 2007; Tucker 2007; Geraldes et al. 2008), retained ancestral polymorphism and introgressive hybridization appear to be sufficiently common that shared polymorphism can often be explained without invoking balancing selection.
Functional properties of BALB/c and MSM/s hemoglobins:
We detected no differences in the O2 equilibria of hemolysates from BALB/c (containing the dminor isoHb) and MSM/s (containing the pminor isoHb) under physiological conditions. This indicates that structural differences in the products of dminor and pminor do not have any significant effects on blood–O2 transport. As shown in Figure 4 and Table 4, Hbs from BALB/c and MSM/s showed no physiologically significant differences in O2 affinity (P50) or cooperativity (n50) under identical buffer conditions. As previously observed for Hbs of the deer mouse, Peromyscus maniculatus (Storz et al. 2009), Hb–O2 affinity was not markedly reduced in the presence of DPG alone (ΔlogP50(DPG-stripped) = 0.15 and 0.13 in BALB/c and MSM/s, respectively; Table 4). The magnitude of the DPG effect in the house mouse Hbs was similar to that observed in lowland deer mice (ΔlogP50(DPG-stripped) = 0.09–0.14), and is indicative of weak DPG binding to the tense-state deoxyHb structure. In contrast to the relatively weak DPG effect, the effect of Cl− ions was much more pronounced (ΔlogP50(KCl-stripped) = 0.29 and 0.31 in BALB/c and MSM/s, respectively; Table 4), which may be attributable to the presence of one or more distinct Cl− binding sites in the tense-state quaternary structure.
Reconciling the results of neutrality tests and functional tests:
A number of studies have documented patterns of DNA sequence variation that provide suggestive evidence for the maintenance of protein polymorphism by balancing selection (e.g., Filatov and Charlesworth 1999; Cork and Purugganan 2002; Tian et al. 2002; Baysal et al. 2007; Ferguson et al. 2008; Fumagalli et al. 2009). However, there are very few case studies where indirect, statistical evidence for balancing selection has been combined with documented functional differences between alternative alleles (for one notable exception involving house mice, see Johnsen et al. 2009). Our combined evolutionary and functional analysis of the two-locus β-globin polymorphism in Indian house mice revealed seemingly contradictory results. The population genetic analysis revealed excess levels of polymorphism at HBB-T2 that could not be reconciled with the expectations of a neutral equilibrium model, and yet the analysis of protein function revealed no discernible physiological differences between dminor and pminor isoHbs. There is clearly no basis for fitness variation among genotypes if the products of alternative alleles are functionally identical.
One possible explanation for the discrepancy is that results of the HKA test reflect a violation of model assumptions related to demographic history or population structure. Since the ratio of intraspecific polymorphism to between-species divergence can be inflated by population structure (Wakeley 2000), results of the HKA test may be biased if the test is applied to a set of loci that are characterized by different levels of population structure (Ingvarsson 2004). This may be a common problem in studies of house mice, since natural populations often show evidence of historical admixture and levels of introgression are highly variable among different genomic regions (e.g., Payseur et al. 2004; Geraldes et al. 2008; Teeter et al. 2008, 2010). Another possibility is that the elevated polymorphism that we observe at the β-globin genes reflects the effects of diversity-enhancing selection at one or more linked loci. We cannot rule out this possibility, but given the observed decay of intragenic LD at each of the two β-globin genes (Figure 3), it does not seem plausible that levels of diversity could be affected by associative overdominance or some other form of diversity-enhancing selection at a linked locus. If the unusual patterns of variation at the β-globin genes of Indian house mice can be explained by admixture or other complexities of population structure, then we should expect that surveys of additional autosomal genes will uncover similar examples of allelic dimorphism and allele sharing between M. castaneus and other closely related species.
Finally, it is also possible that the β-globin polymorphism in M. castaneus is maintained by balancing selection, but that the adaptive variation in protein function is not directly related to blood–O2 transport. In contrast to the allelic differences in cysteine content that distinguish products of Hbbd and Hbbs in M. domesticus and M. musculus, amino acid differences between the dminor and pminor isoHbs of M. castaneus have no effect on the metabolism of thiol reactants or intraerythrocytic redox balance (which can affect aspects of the host immune response to pathogenic infection). Nonetheless, we cannot rule out the possibility that the β-globin polymorphism in M. castaneus affects some unknown biochemical function, especially in light of recent discoveries concerning the expression of globin chain monomers in nonerythroid cells (e.g., Liu et al. 1999; Mansergh et al. 2008; Nishi et al. 2008; Biagioli et al. 2009; Richter et al. 2009; Schelshorn et al. 2009). In light of our experimental results, we conclude that amino acid differences between products of the dminor and pminor alleles are functionally inconsequential with regard to the respiratory physiology of house mice. If the dminor and pminor alleles are maintained as a balanced polymorphism in M. castaneus, either directly or indirectly, the associated variance in fitness appears to be unrelated to respiratory functions of Hb.
Acknowledgments
We thank B. Harr for sharing tissue samples, and we thank A. Geraldes and M. Nachman for shipping samples and sharing sequence data. We also thank M.-B. Hemmingsen for valuable assistance in the lab. Finally, we are grateful to A. Geraldes, M. Nachman, and two anonymous reviewers for helpful comments that improved the manuscript. This work was funded by a National Science Foundation fellowship in Bioinformatics to A.M.R. (0630779), grants to J.F.S. from the National Science Foundation and the Nebraska Research Council, and grants to J.F.S., R.E.W., and A.F. from the National Institutes of Health/National Heart, Lung, and Blood Institute.
References
- Baines, J. F., and B. Harr, 2007. Reduced X-linked diversity in derived populations of house mice. Genetics 175 1911–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal, B., E. Lawrence and R. Ferrell, 2007. Sequence variation in human succinate dehydrogenase genes: evidence for long-term balancing selection on SDHA. BMC Biol. 5 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, R., and M. Jakobson, 1975. Ecological genetics of an island population of the house mouse. J. Zool. 173 341–354. [Google Scholar]
- Berry, R., and H. Murphy, 1970. The biochemical genetics of an island population of the house mouse. Proc. R. Soc. Lond. Ser. B 176 87–103. [Google Scholar]
- Berry, R., and J. Peters, 1975. Macquarie Island house mice: a genetical isolate on a sub-Antarctic island. J. Zool. 176 375–389. [Google Scholar]
- Berry, R., and J. Peters, 1977. Heterogeneous heterozygosities in Mus musculus populations. Proc. R. Soc. Lond. Ser. B 197 485–503. [DOI] [PubMed] [Google Scholar]
- Berry, R., and J. Peters, 1981. Allozymic variation in house mouse populations, pp. 242–253 in Mammalian Population Genetics, edited by M. Smith and J. Joule. University of Georgia Press, Athens, Georgia.
- Berry, R., J. Peters and R. Van Aarde, 1978. Sub-antarctic house mice: colonization, survival, and selection. J. Zool. 184 127–141. [Google Scholar]
- Berry, R. J., 1978. Genetic variation in wild house mice: where natural selection and history meet. Am. Sci. 66 52–60. [PubMed] [Google Scholar]
- Betrán, E., J. Rozas, A. Navarro and A. Barbadilla, 1997. The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics 146 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagioli, M., M. Pinto, D. Cesselli, M. Zaninello, D. Lazarevic et al., 2009. Unexpected expression of alpha- and beta-globin in mesencephalic dopaminergic neurons and glial cells. Proc. Natl. Acad. Sci. USA 106 15454–15459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme, F., N. Miyashita, P. Boursot, J. Catalan and K. Moriwaki, 1989. Genetical variation and polyphyletic origin in Japanese Mus musculus. Heredity 63 299–308. [DOI] [PubMed] [Google Scholar]
- Boursot, P., J. Auffray, J. Britton-Davidian and F. Bonhomme, 1993. The evolution of house mice. Ann. Rev. Ecol. Syst. 24 119–152. [Google Scholar]
- Boursot, P., W. Din, R. Anand, D. Darviche, B. Dod et al., 1996. Origin and radiation of the house mouse: mitochondrial DNA phylogeny. J. Evol. Biol. 9 391–415. [Google Scholar]
- Charlesworth, B., D. Charlesworth and N. Barton, 2003. The effects of genetic and geographic structure on neutral varation. Ann. Rev. Ecol. Evol. Syst. 34 99–125. [Google Scholar]
- Charlesworth, D., 2006. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2 e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cork, J., and M. Purugganan, 2002. High-diversity genes in the Arabidopsis genome. Genetics 170 1897–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhart, M. A., K. S. Simons and S. Weaver, 1985. Evolution of the mouse β-globin gene: a recent gene conversion in the Hbbs haplotypes. Mol. Biol. Evol. 2 304–320. [DOI] [PubMed] [Google Scholar]
- Ferguson, W., S. Dvora, J. Gallo, A. Orth and S. Boissinot, 2008. Long-term balancing selection at the West-Nile virus resistance gene, Oas1b, maintains trans-specific polymorphisms in the house mouse. Mol. Biol. Evol. 25 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov, D., and D. Charlesworth, 1999. DNA polymorphism, haplotype structure and balancing selection in the Leavenworthia PgiC locus. Genetics 153 1423–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli, M., R. Cagliani, U. Pozzoli, S. Riva, G. P. Comi et al., 2009. Widespread balancing selection and pathogen-driven selection at blood group antigen genes. Genome Res. 19 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes, A., Basset, B. Bigson, K. Smith, B. Harr et al., 2008. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked, and mitochondrial genes. Mol. Ecol. 17 5349–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman, J. G., 1972. Hemoglobin β-chain structural variation in mice: evolutionary and functional implications. Science 178 873–874. [DOI] [PubMed] [Google Scholar]
- Gilman, J. G., 1974. Rodent hemoglobin structure: a comparison of several species of mice. Ann. N. Y. Acad. Sci. 241 416–433. [DOI] [PubMed] [Google Scholar]
- Giustarini, D., I. Dalle-Donne, E. Cavarra, S. Fineschi, G. Lungarella et al., 2006. Metabolism of oxidants by blood from different mouse strains. Biochem. Pharmacol. 71 1753–1764. [DOI] [PubMed] [Google Scholar]
- Guo, S., and E. Thompson, 1992. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48 361–372. [PubMed] [Google Scholar]
- Hayashi, A., T. Suzuki and M. Shin, 1973. An enzymatic reduction system for metmyoglobin and methemoglobin, and its application to functional studies of oxygen carriers. Biochim. Biophys. Acta 310 309–316. [DOI] [PubMed] [Google Scholar]
- Hempe, J. M., J. Ory-Ascani and D. Hsia, 2007. Genetic variation in mouse beta globin cysteine content modifies glutathione metabolism: implications for the use of mouse models. Exp. Biol. Med. 232 437–444. [PubMed] [Google Scholar]
- Hey, J., and J. Wakeley, 1997. A coalescent estimator of the population recombination rate. Genetics 145 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, W. G., and B. Weir, 1988. Variances and covariances of squared linkage disequilibria in finite populations. Theor. Popul. Biol. 33 54–78. [DOI] [PubMed] [Google Scholar]
- Hoffmann, F. G., J. C. Opazo and J. F. Storz, 2008. New genes originated via multiple recombinational pathways in the β-globin gene family of rodents. Mol. Biol. Evol. 25 2589–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R., and N. Kaplan, 1988. The coalescent process in models with selection and recombination. Genetics 120 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton, J., J. Bishop, R. Schweet and E. Russell, 1962. Hemoglobin inheritance in inbred mouse strains, II. Genetic studies. Proc. Natl. Acad. Sci. USA 48 1718–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson, P. K., 2004. Population subdivision and the Hudson-Kreitman-Aguade test: testing for deviations from the neutral model in organelle genomes. Genet. Res. 83 31–39. [DOI] [PubMed] [Google Scholar]
- Johnsen, J. M., M. Teschke, P. Pavlidis, B. M. McGee, D. Tautz et al., 2009. Selection on cis-regulatory variation at B4galnt2 and its influence on von Willebrand factor in house mice. Mol. Biol. Evol. 26 567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima, T., N. Miyashita, C.-H. Wang, X.-Q. He, M.-L. Jin et al., 1991. A new haplotype of the β-globin gene complex, Hbbw1, in Chinese wild mice. Jpn. J. Genet. 66 491–500. [DOI] [PubMed] [Google Scholar]
- Kawashima, T., N. Miyashita, K. Tsuchiya, H. Li, F. S. Wang et al., 1995. Geographical-distribution of the Hbb haplotypes in the Mus musculus subspecies in Eastern Asia. Jpn. J. Genet. 70 17–23. [DOI] [PubMed] [Google Scholar]
- Kelly, J. K., 1997. A test of neutrality based on interlocus associations. Genetics 146 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado, P., and J. Rozas, 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 1451–1452. [DOI] [PubMed] [Google Scholar]
- Liu, L., M. Zeng and J. S. Stamler, 1999. Hemoglobin induction in mouse macrophages. Proc. Natl. Acad. Sci. USA 96 6643–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansergh, F. C., S. M. Hunter, J. C. Geatrell, M. Jarrin, K. Powell et al., 2008. Developmentally regulated expression of hemoglobin subunits in avascular tissues. Int. J. Dev. Biol. 52 873–886. [DOI] [PubMed] [Google Scholar]
- Minezawa, M., K. Moriwaki and K. Kondo, 1979. Geographical distribution of Hbbp allele in the Japanese wild mouse, Mus musculus molossinus. Jpn. J. Genet. 54 166–173. [Google Scholar]
- Miranda, J., 2000. Highly reactive cysteine residues in rodent hemoglobins. Biochem. Biophys. Res. Commun. 275 517–523. [DOI] [PubMed] [Google Scholar]
- Miyashita, N., K. Moriwaki, M. Minezawa, H. Yonekawa, F. Bonhomme et al., 1985. Allelic constitution of the hemoglobin β chain in wild populations of the house mouse, Mus musculus. Biochem. Genet. 23 975–986. [DOI] [PubMed] [Google Scholar]
- Myers, J., 1974. Genetic and social structure of feral house mouse populations on Grizzly Island, California. Ecology 55 747–759. [Google Scholar]
- Nei, M., and S. Kumar, 2000. Molecular Evolution and Phylogenetics. Oxford University Press, New York.
- Nishi, H., R. Inagi, H. Kato, M. Tanemoto, I. Kojima et al., 2008. Hemoglobin is expressed by mesangial cells and reduces oxidant stress. J. Am. Soc. Nephrol. 19 1500–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth, A., T. Adama, W. Din and F. Bonhomme, 1998. Hybridation naturelle entre deux sous-espèces de souris domestique, Mus musculus domesticus et Mus musculus castaneus, près du lac Casitas (Californie). Genome 41 104–110. [DOI] [PubMed] [Google Scholar]
- Payseur, B. A., J. G. Krenz and M. W. Nachman, 2004. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58 2064–2078. [DOI] [PubMed] [Google Scholar]
- Petras, M., 1967. Studies of natural populations of Mus. I. Biochemical polymorphisms and their bearing on breeding structure. Evolution 21 259–274. [DOI] [PubMed] [Google Scholar]
- Petras, M., and J. Topping, 1983. The maintenance of polymorphisms at two loci in house mouse (Mus musculus) populations. Can. J. Genet. Cytol. 25 190–201. [DOI] [PubMed] [Google Scholar]
- Polzin, T., and S. Daneschmand, 2003. On Steiner trees and minimum spanning trees in hypergraphs. Oper. Res. Lett. 31 12–20. [Google Scholar]
- Richter, F., B. H. Meurers, C. N. Zhu, V. P. Medvedeva and M. F. Chesselet, 2009. Neurons express hemoglobin alpha- and beta-chains in rat and human brains. J. Comp. Neurol. 515 538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte, U., and E. Neufeld, 1982. East Asian hemoglobin type (Hbbp) in wild populations of the house mouse in Israel. Biochem. Genet. 5/6 475–481. [DOI] [PubMed] [Google Scholar]
- Rozas, J., M. Gullaud, G. Blandin and M. Aguadé, 2001. DNA variation at the rp49 gene region of Drosophila simulans: evolutionary inferences from an unusual haplotype structure. Genetics 158 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runck, A. M., H. Moriyama and J. F. Storz, 2009. Evolution of duplicated β-globin genes and the structural basis of hemoglobin isoform differentiation in Mus. Mol. Biol. Evol. 26 2521–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage, R., 1981. Wild mice, pp. 39–90 in The Mouse in Biomedical Research: History, Genetics, and Wild Mice, edited by H. Foster, J. Small and J. Fox. Academic Press, New York.
- Sage, R., J. B. I. Whitney and A. Wilson, 1986. Genetic analysis of a hybrid zone between domesticus and musculus mice (Mus musculus complex): hemoglobin polymorphisms. Curr. Top. Microbiol. 127 75–85. [DOI] [PubMed] [Google Scholar]
- Salcedo, T., A. Geraldes and M. W. Nachman, 2007. Nucleotide variation in wild and inbred mice. Genetics 177 2277–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, J. J., A. Shinohara, N. Miyashita, C. Koshimoto, K. Tsuchiya et al., 2008. Discovery of a new HBB haplotype w2 in a wild-derived house mouse, Mus musculus. Mamm. Genome 19 155–162. [DOI] [PubMed] [Google Scholar]
- Schelshorn, D. W., A. Schneider, W. Kuschinsky, D. Weber, C. Kruger et al., 2009. Expression of hemoglobin in rodent neurons. J. Cerebr. Blood F. Met. 29 585–595. [DOI] [PubMed] [Google Scholar]
- Selander, R., and S. Yang, 1969. Protein polymorphism and genic heterozygosity in a wild population of the house mouse. Genetics 63 653–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander, R., W. Hunt and S. Yang, 1969. a Protein polymorphism and genic heterozygosity in two European subspecies of the house mouse. Evolution 23 379–390. [DOI] [PubMed] [Google Scholar]
- Selander, R., S. Yang and W. Hunt, 1969. b Polymorphisms in esterases and hemoglobin in wild populations of the house mouse (Mus musculus). Univ. Texas. Publ. 6918 271–338. [Google Scholar]
- She, J., F. Bonhomme, P. Boursot, L. Thaler and F. Catzeflis, 1990. Molecular phylogenies in the genus Mus: comparative analysis of electrophoretic, scnDNA hybridization, and mtDNA RFLP data. Biol. J. Linn. Soc. 41 83–103. [Google Scholar]
- Storz, J. F., and H. Moriyama, 2008. Mechanisms of hemoglobin adaptation to high-altitude hypoxia. High Alt. Med. Biol. 9 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz, J. F., M. Baze, J. Waite, F. G. Hoffmann, J. C. Opazo et al., 2007. Complex signatures of selection and gene conversion in the duplicated globin genes of house mice. Genetics 177 481–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz, J. F., A. M. Runck, S. J. Sabatino, J. K. Kelly, N. Ferrand et al., 2009. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc. Natl. Acad. Sci. USA 106 14450–14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., T. Shimada, M. Terashima, K. Tsuchiya and K. Aplin, 2004. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol. Phylogenet. Evol. 33 626–646. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 122 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter, K. C., B. A. Payseur, L. W. Harris, M. A. Bakewell, L. M. Thibodeau et al., 2008. Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Res. 18 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter, K. C., L. M. Thibodeau, Z. Gompert, C. A. Buerkle, M. W. Nachman et al., 2010. The variable genomic architecture of isolation between hybridizing species of house mouse. Evolution 54 472–485. [DOI] [PubMed] [Google Scholar]
- Thompson, J., T. Gibso, F. Plewniak, F. Jeanmougin and D. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D., H. Araki, E. Stahl, J. Bergelson and M. Kreitman, 2002. Signature of balancing selection in Arabadopsis. Proc. Natl. Acad. Sci. USA 99 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, P. K., 2007. Systematics of the genus Mus, pp. 13–23 in The Mouse in Biochemical Research, edited by J. Fox, C. Newcomer, A. Smith, S. Barthold, F. Quimby et al. Elsevier Press, Boston.
- Ueda, Y., N. Miyashita, K. Imai, Y. Yamaguchi, K. Takamura et al., 1999. Nucleotide sequences of the mouse globin β gene cDNAs in a wild derived new haplotype Hbbw1. Mamm. Genome 10 879–882. [DOI] [PubMed] [Google Scholar]
- Wakeley, J., 2000. The effects of subdivision on the genetic divergence of populations and species. Evolution 54 1092–1101. [DOI] [PubMed] [Google Scholar]
- Watterson, E., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7 256–276. [DOI] [PubMed] [Google Scholar]
- Weber, R. E., 1981. Cationic control of oxygen affinity in lungworm erythocruorin. Nature 292 386–387. [Google Scholar]
- Weber, R. E., 1992. Use of ionic and zwitterionic (Tris/BisTris and Hepes) buffers in studies on hemoglobin function. J. Appl. Physiol. 72 1611–1615. [DOI] [PubMed] [Google Scholar]
- Weber, R. E., and A. Fago, 2004. Functional adaptation and its molecular basis in vertebrate hemoglobins, neuroglobins and cytoglobins. Resp. Physiol. Neurobi. 144 141–159. [DOI] [PubMed] [Google Scholar]
- Weber, R. E., W. Voelter, A. Fago, H. Echner, E. Campanella et al., 2004. Modulation of red cell glycolysis: interaction between vertebrate hemoglobins and cytoplasmic domains of band 3 red cell membrane proteins. Am. J. Physiol. 287 R454–R464. [DOI] [PubMed] [Google Scholar]
- Weir, B., and C. Cockerham, 1984. Estimating F-statistics for the analysis of population structure. Evolution 38 1358–1370. [DOI] [PubMed] [Google Scholar]
- Wheeler, L., and R. Selander, 1972. Genetic variation in populations of the house mouse, Mus musculus, in the Hawaiian Islands. Univ. Texas Publ. 7213 269–296. [Google Scholar]
- Whitney, J. B. I., 1977. Differential control of the synthesis of two hemoglobin beta chains in normal mice. Cell 12 863–871. [DOI] [PubMed] [Google Scholar]