Figure 7.
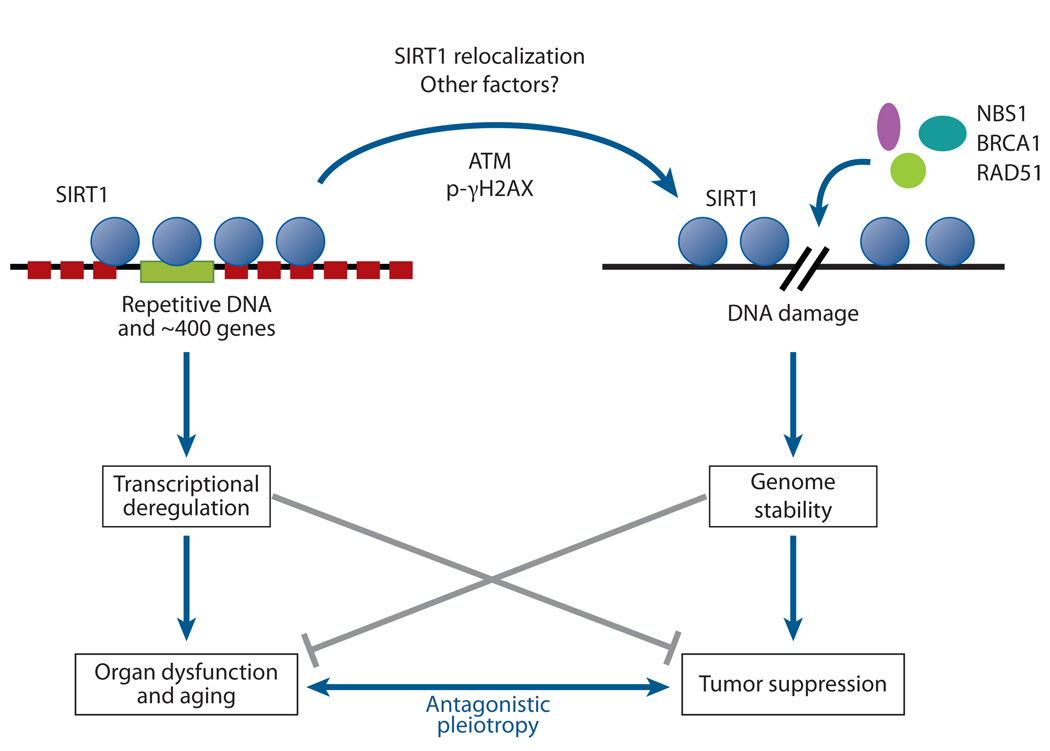
The relocalization of chromatin modifiers (RCM) hypothesis of aging stems from S. Imai and H. Kitano, who proposed in 1998 that changes in heterochromatin underlie the aging process. The idea was based in part on observations that, in response to DNA damage and aging, yeast SIR2 is released from silent loci and relocalized to DNA breaks, where it is hypothesized to organize chromatin to facilitate repair. During relocalization, expression of silent mating-type genes (HML and HMR) cause sterility, a hallmark of aging. Recent work shows that a similar process may drive aging in mammals. In response to DNA breaks or aging, SIRT1 also relocalizes away from open reading frames (ORFs) to DNA-break sites, seemingly to alter chromatin around the break site and recruit DNA damage–repair proteins such as RAD51 and NBS1. This relocalization of SIRT1, or the epigenetic changes it induces, is proposed to alter gene-expression patterns that result in tissue dysfunction and diseases associated with aging.