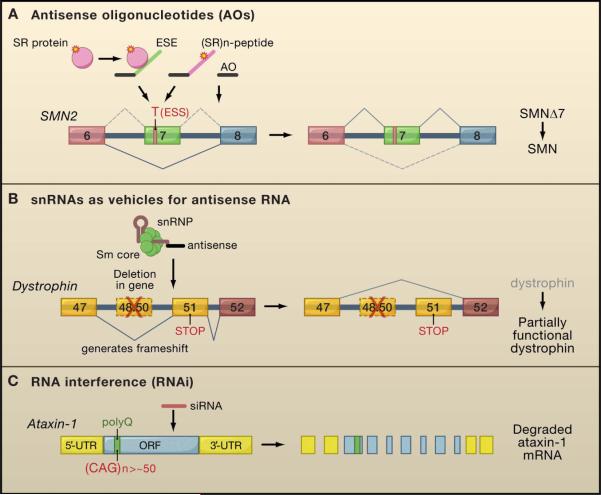
Figure 4. Therapeutic approaches that use or target RNAs.
A. Antisense oligonucleotides (AOs) or bifunctional AOs that have sequences complementary to exon 7 conjugated to splicing-enhancing effectors---e.g., serine-arginine (SR) peptide or an ESE that recruits SR proteins---are used to promote exon 7 inclusion of the SMN2 gene, decreasing the production of a truncated SMN protein (SMNΔ7) and increasing that of a full-length functional protein (SMN).
B. Modified U1 or U7 snRNA-based vectors can carry complementary sequences to the targeting RNAs to achieve the desired splicing pattern. As shown here, genomic deletions (e.g., from exon 48 to 50) in dystrophin genes of DMD patients lead to a premature termination codon (STOP) in exon 51 caused by a frameshift. Antisense U7 snRNA targeting the 3′ splice site of exon 51 prevents the inclusion of this exon and restores the reading frame, and as a result, produces a partially functional protein.
C. RNAi approaches are used to eliminate pathogenic mRNAs. siRNAs delivered to a mouse model of spinocerebellar ataxia type I (SCA1) elicit degradation and efficient knockdown of the disease-causing ataxin-1 mRNA that contains the expanded CAG repeats encoding polyglutamine (polyQ).