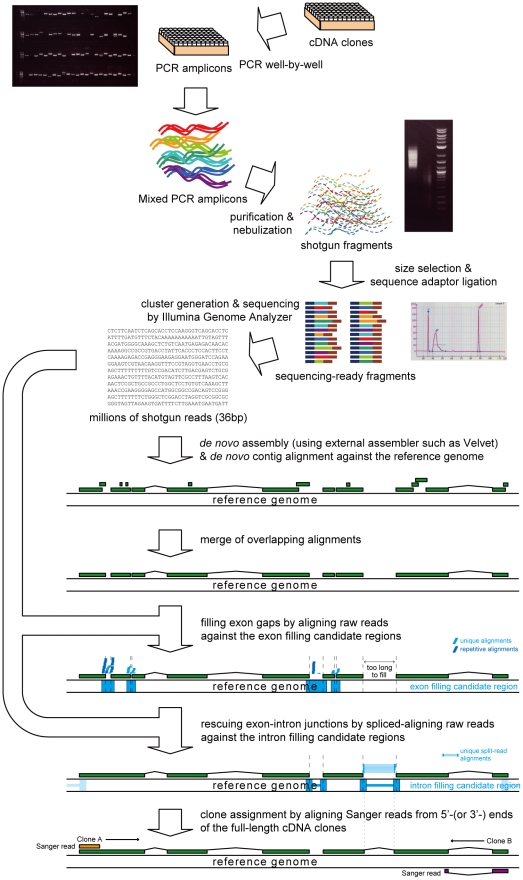
Figure 1. Whole full-length cDNA clone sequencing process using MuSICA 2.
Target cDNA clones are prepared (for example, 800 clones per flow cell). The clones are individually amplified by PCR. The amplicons are mixed in equal volumes, and then fragmented by nebulization. The fragments of appropriate size for sequencing are size-fractionated by gel electrophoresis, followed by sequence adaptor ligation. A number of 36-bp reads are collected using the Illumina GA. The obtained reads are assembled by an external de novo assembler such as Velvet. The assembled contigs are then aligned against the reference genome sequence to merge overlapping contigs. Missing exons are filled by aligning raw reads against small gaps between the merged contigs. Missing introns (links between exons) are recovered by spliced alignment of the raw reads. The final contigs are associated with individual full-length cDNA clones by Sanger reads from either/both ends of the clones.