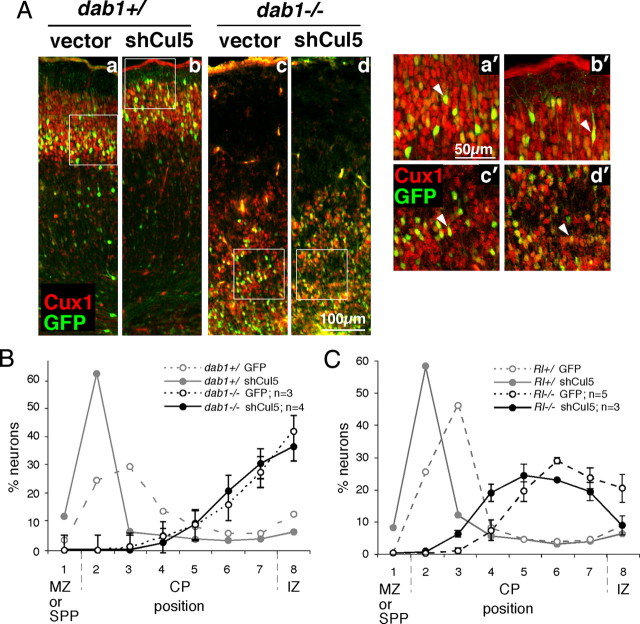
Figure 2.
Cul5 requires Dab1 to regulate neuron position. A, Embryos resulting from a dab1+/− mating were subject to in utero microinjection and electroporation at E14.5 with 2 μg of pCAG-GFP+MFE (vector, a and c) or pCAG-GFP-shCul5 (shCul5, b and d). Embryos were genotyped and sections were prepared at E19.5 and stained with Cux1 antibody (red), which stains neurons generated between approximately E14.5 and E17.5. Cux1 staining indicates the inversion of the cortical layering in the dab1−/− mutants (c and d) versus the control wild-type or heterozygous embryos (a and b). GFP-positive neurons migrate approximately with their cohort Cux1-positive sisters in all cases. However, in control embryos, shCul5 induces a shift upward in neuron position (b) compared with control plasmid (a). In contrast, in dab1−/− embryos, in utero microinjection of the same quantities of shCul5 (d) and control plasmid (c) did not modify neuron position. (a′–d′) Single-plane high-power images taken with a confocal microscope showed that in all conditions GFP-positive neurons continue to express Cux1, indicating that their fate is unchanged. B, Neuron positions at E19.5 after Cul5 depletion in dab1−/− embryos, as shown in A. Results are mean ± SEM. There is no significant difference between control and shCul5 neurons in dab1−/− embryos. SPP indicates superplate. C, Neuron positions at E19.5 after Cul5 depletion in Rl−/− embryos, as shown in A but using embryos from a Rl+/− mating. Results are mean ± SEM. Cul5 shRNA causes a significant difference in mean neuron position in Rl−/− embryos (control, 6.21 ± 0.26, n = 5; shCul5, 5.55 ± 0.13, n = 3; p = 0.017).