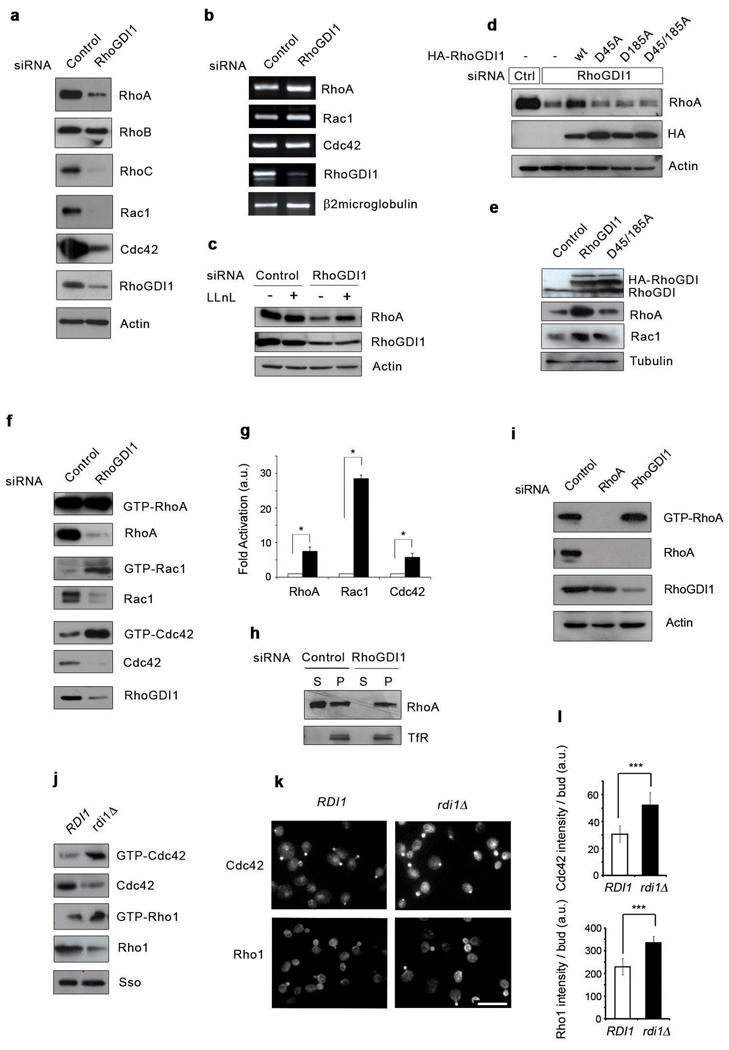
Figure 1. RhoGDI1depletion triggers both degradation and activation of Rho proteins in eukaryotic cells.
(a) Lysates from control or RhoGDI1 siRNA transfected HeLa cells were resolved by SDS-PAGE and analyzed by Western blotting. (b) Total RNAs were purified from control or RhoGDI1 siRNA transfected cells. RT-PCR was performed on DNAse I treated RNA using specific primers. RT-PCR products were resolved by agarose gel electrophoresis. (c) Lysates from control or RhoGDI1 siRNA transfected HeLa cells treated with LLnL for 8 hours were resolved by SDS-PAGE and analyzed by Western blotting. (d) HeLa cells were co-transfected with control or RhoGDI1 siRNA and HA-tagged RhoGDI1 RNAi resistant mutants. Cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. All results are representative of at least 3 independent experiments. Note that the degradation of RhoA in RhoGDI1-depleted cells is rescued by expression of RhoGDI1 resistant to the siRNA, but not by RhoGDI1 mutants that do not bind Rho GTPases. (e) Lysates from HeLa cells transfected with HA-tagged wt RhoGDI1 or RhoGDI1 D45/185A were resolved by SDS-PAGE and analyzed by Western blotting. All siRNA experiments were analyzed at 72 h after transfection. For overexpression experiments, cells were transfected with the indicated cDNA 48 h after siRNA transfection and incubated for an additional 24 h. (f) HeLa cells were transfected with control or RhoGDI1 siRNA for 72 h. Active Rho GTPases were pulled-down from cell lysates with GST-RBD or GST-PBD beads. Bound proteins and total cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. (g) Quantitation of the activation of RhoA, Rac1 and Cdc42 in RhoGDI1-depleted cells (black bars) relative to control cells (white bars). * p=0.0108, p=0.0146 and p=0.0229 respectively for RhoA, Rac1 and Cdc42 in a two-tail unpaired student’s t test with error bars representing standard error of the mean (s.e.m.) from three independent experiments. (h) Control or RhoGDI1 siRNA transfected cells were fractionated into cytosol and membrane fractions, resolved by SDS-PAGE and analyzed by Western blotting. TfR stands for transferrin receptor. (i) HeLa cells were infected with a RhoA miR shRNA-expressing adenovirus or transfected with RhoGDI1 siRNA for 72 h. Active RhoA was pulled-down with GST-RBD. Total cell lysates and GST-RBD bound proteins were resolved by SDS-PAGE and analyzed by Western blotting. (j) Active Rho GTPases from wild-type or Rdi1d yeast strains were pulled-down with GTP-RBD or GST-PBD beads. Bound proteins and total cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. The Sso protein is used as loading control. (k) Immunofluorescence staining of Cdc42 and Rho1 in control wild-type (RDI1) and rdi1Δ strains. Scale bar is 8µm. (l) Quantitation of the intensity of polarized bud staining in control wild-type (RDI1) and rdi1Δ strains. Data were analyzed by two-tail student’s t test (p < 0.005) with error bars representing standard deviation from three experiments.