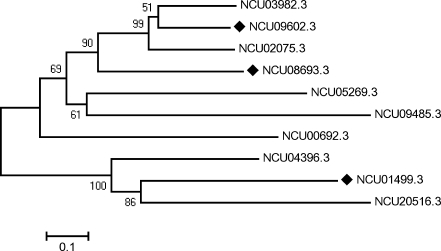
Fig. 1.
Phylogenetic analysis of Neurospora crassa hsp70 genes. Multiple alignments of 10 HSP70 amino acid sequences using the N. crassa genome database was performed using ClustalW (http://www.ebi.ac.uk/clustalw). A phylogenetic tree was constructed from these alignments using the neighbor-joining method with the software package MEGA version 3.1 (http://www.megasoftware.net). Three of them were chosen for transcription analysis (filled diamonds). The number at the nodes represents the percentage of bootstrap values (1,000 replicates). The scale bar represents the phylogenetic distance of 0.1 amino acid substitution per site