Fig. 3.
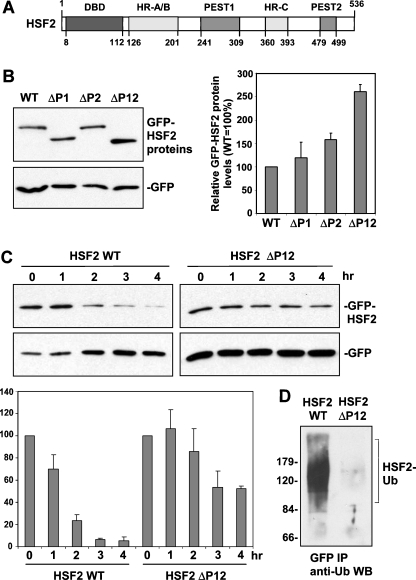
Role of two PEST domains in HSF2 instability. a Schematic showing location of two PEST domains in HSF2 relative to the DNA-binding domain (DBD), Heptad Repeat oligomerization domain (HR-A/B), and Heptad Repeat C (HR-C). b Extracts of cells transfected with GFP fusion constructs of wild type, PEST domain 1-deleted (ΔP1), PEST domain 2-deleted (ΔP2), and PEST domain 1- and 2-deleted (ΔP12) HSF2, along with GFP alone expression plasmid (normalizing control) were subjected to Western blot using anti-GFP antibodies. For the graph shown to the right of this data, bands of the different GFP-HSF2 fusion proteins from replicates of this experiment were quantified, normalized to values of the GFP alone bands, and then graphed as percent relative to that of wild-type HSF2 (which was set to 100%). Data are presented as means ±SEM (n = 3). c HeLa cells were transfected with the GFP-HSF2 wild-type or the GFP-HSF2 construct lacking the PEST domains along with GFP alone expression plasmid (normalizing control), treated with cycloheximide to block translation, and then extracts of cells collected at the indicated time points after addition of cycloheximide were subjected to anti-GFP Western blot. For the graph shown below this data, bands of GFP-HSF2 fusion proteins from replicates of this experiment were quantified, normalized to values of GFP bands, and graphed as percent of wild-type HSF2 in the absence of cycloheximide (set to 100%). Data are presented as means ± SEM (n = 3). d Cells transfected with GFP-HSF2 wild-type and GFP-HSF2 lacking the PEST domains were treated with MG132 for 4 h, and then extracts of these cells were subjected to anti-GFP immunoprecipitation followed by anti-ubiquitin Western blot assay. Positions of molecular weight markers are indicated to the left of the panel