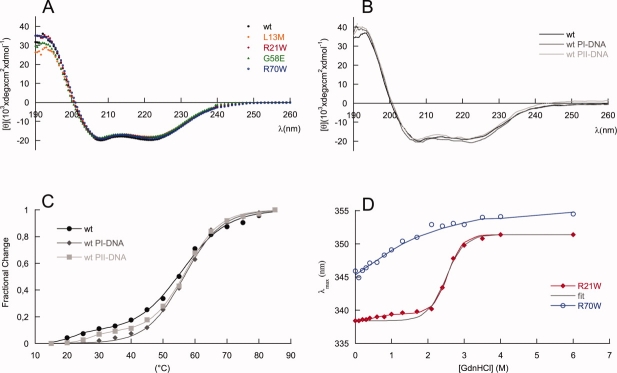
Figure 4.
MexR structural effects of mutations and DNA binding. Far UV CD spectra of (A) MexR-wt (filled circle, black), MexR-L13M (empty rhomb, orange), MexR-R21W (filled rhomb, red), MexR-G58E (filled triangle, green), MexR-R70W (empty circle, blue) and (B) MexR-wt (solid black line) in complex with DNA binding boxes PI (solid line, dark grey) and PII (solid line, light grey) show that the structural effects of the studied MDR mutations are negligible. C: Thermal denaturation measurements at 222 nm for MexR-wt (filled circle, black), MexR-wt with PI-DNA (filled rhomb, dark grey), and MexR-wt with PII-DNA (filled square, light grey) indicate a less stable DNA-binding domain that is stabilized on DNA binding. The corresponding fits for two- and three-state denaturation (Table 4) are shown as solid lines. D: Gdn-HCl induced unfolding of MexR R21W (filled rombs, red) and R70W (empty circles, blue) indicate that the most stable MexR region is located in the dimer interface, while the DNA-binding region is unstable. Two-state (black) and three-state (red) fits (Table 4) to R21W data are shown. The R70W data were not sufficient for a full thermodynamic fit, thus the blue line only represents a graphic best fit.